This is a preprint.
Protein folding as a jamming transition
- PMID: 38800654
- PMCID: PMC11118678
Protein folding as a jamming transition
Abstract
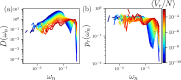
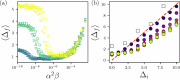
Proteins fold to a specific functional conformation with a densely packed hydrophobic core that controls their stability. We develop a geometric, yet all-atom model for proteins that explains the universal core packing fraction of found in experimental measurements. We show that as the hydrophobic interactions increase relative to the temperature, a novel jamming transition occurs when the core packing fraction exceeds . The model also recapitulates the global structure of proteins since it can accurately refold to native-like structures from partially unfolded states.
Figures


Similar articles
-
Desolvation and development of specific hydrophobic core packing during Im7 folding.J Mol Biol. 2010 Mar 12;396(5):1329-45. doi: 10.1016/j.jmb.2009.12.048. Epub 2010 Jan 4. J Mol Biol. 2010. PMID: 20053361 Free PMC article.
-
Shear thickening and jamming in densely packed suspensions of different particle shapes.Phys Rev E Stat Nonlin Soft Matter Phys. 2011 Sep;84(3 Pt 1):031408. doi: 10.1103/PhysRevE.84.031408. Epub 2011 Sep 28. Phys Rev E Stat Nonlin Soft Matter Phys. 2011. PMID: 22060372
-
Side chain burial and hydrophobic core packing in protein folding transition states.Protein Sci. 2008 Apr;17(4):644-51. doi: 10.1110/ps.073105408. Epub 2008 Feb 27. Protein Sci. 2008. PMID: 18305200 Free PMC article.
-
Pathways of protein folding.Annu Rev Biochem. 1993;62:653-83. doi: 10.1146/annurev.bi.62.070193.003253. Annu Rev Biochem. 1993. PMID: 8352599 Review.
-
Toward a better understanding of protein folding pathways.Proc Natl Acad Sci U S A. 1988 Jul;85(14):5082-6. doi: 10.1073/pnas.85.14.5082. Proc Natl Acad Sci U S A. 1988. PMID: 2455892 Free PMC article. Review.
References
-
- Dill K. A., Dominant forces in protein folding, Biochemistry 29, 7133 (1990). - PubMed
-
- Wolynes P. G., Onuchic J. N., and Thirumalai D., Navigating the folding routes, Science 267, 1619 (1995). - PubMed
-
- Onuchic J. N., Luthey-Schulten Z., and Wolynes P. G., Theory of protein folding: The energy landscape perspective, Annual Review of Physical Chemistry 48, 545 (1997). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
