Parsing the roles of DExD-box proteins DDX39A and DDX39B in alternative RNA splicing
- PMID: 38801080
- PMCID: PMC11317157
- DOI: 10.1093/nar/gkae431
Parsing the roles of DExD-box proteins DDX39A and DDX39B in alternative RNA splicing
Abstract
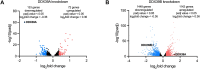
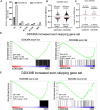
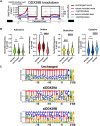
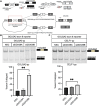
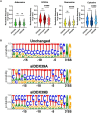
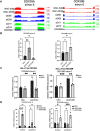
DExD-box RNA proteins DDX39A and DDX39B are highly homologous paralogs that are conserved in vertebrates. They are required for energy-driven reactions involved in RNA processing. Although we have some understanding of how their functions overlap in RNA nuclear export, our knowledge of whether or not these proteins have specific or redundant functions in RNA splicing is limited. Our previous work has shown that DDX39B is responsible for regulating the splicing of important immune transcripts IL7R and FOXP3. In this study, we aimed to investigate whether DDX39A, a highly homologous paralog of DDX39B, plays a similar role in regulating alternative RNA splicing. We find that DDX39A and DDX39B have significant redundancy in their gene targets, but there are targets that uniquely require one or the other paralog. For instance, DDX39A is incapable of complementing defective splicing of IL7R exon 6 when DDX39B is depleted. This exon and other cassette exons that specifically depend on DDX39B have U-poor/C-rich polypyrimidine tracts in the upstream intron and this variant polypyrimidine tract is required for DDX39B dependency. This study provides evidence that despite a high degree of functional redundancy, DDX39A and DDX39B are selectively required for the splicing of specific pre-mRNAs.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
SNRPB-mediated regulation of DDX39A splicing promotes ovarian cancer progression by regulating α6 integrin subunit expression.Oncogene. 2025 Jul;44(26):2170-2185. doi: 10.1038/s41388-025-03386-0. Epub 2025 Apr 11. Oncogene. 2025. PMID: 40216968
-
Positioning of pyrimidine motifs around cassette exons defines their PTB-dependent splicing in Arabidopsis.Plant J. 2024 Jun;118(6):2202-2218. doi: 10.1111/tpj.16739. Epub 2024 Apr 5. Plant J. 2024. PMID: 38578875
-
De novo and inherited variants in DDX39B cause a novel neurodevelopmental syndrome.Brain. 2025 Aug 1;148(8):2658-2670. doi: 10.1093/brain/awaf035. Brain. 2025. PMID: 39918047
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
Cited by
-
Identification of Novel Modulators of the ALT Pathway Through a Native FISH-Based Optical Screen.bioRxiv [Preprint]. 2024 Nov 15:2024.11.15.623791. doi: 10.1101/2024.11.15.623791. bioRxiv. 2024. Update in: Cell Rep. 2025 Jan 28;44(1):115114. doi: 10.1016/j.celrep.2024.115114. PMID: 39605432 Free PMC article. Updated. Preprint.
-
The role of BUD31 in clear cell renal cell carcinoma: prognostic significance, alternative splicing, and tumor immune environment.Clin Exp Med. 2024 Aug 13;24(1):191. doi: 10.1007/s10238-024-01451-8. Clin Exp Med. 2024. PMID: 39136845 Free PMC article.
-
Chemoproteomic Approach for Identifying Nuclear Arsenite-Binding Proteins.Chem Res Toxicol. 2025 May 19;38(5):954-961. doi: 10.1021/acs.chemrestox.5c00107. Epub 2025 Apr 27. Chem Res Toxicol. 2025. PMID: 40289526
-
Inefficient recruitment of DDX39B impedes pre-spliceosome assembly on FOXP3 introns.RNA. 2024 Jun 17;30(7):824-838. doi: 10.1261/rna.079933.123. RNA. 2024. PMID: 38575347 Free PMC article.
-
TRAIP enhances progression of tongue squamous cell carcinoma through EMT and Wnt/β-catenin signaling by interacting with DDX39A.BMC Cancer. 2024 Dec 2;24(1):1481. doi: 10.1186/s12885-024-13130-8. BMC Cancer. 2024. PMID: 39623306 Free PMC article.
References
-
- Cordin O., Banroques J., Tanner N.K., Linder P.. The DEAD-box protein family of RNA helicases. Gene. 2006; 367:17–37. - PubMed
-
- Linder P., Jankowsky E.. From unwinding to clamping — the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011; 12:505–516. - PubMed
-
- Yamazaki T., Fujiwara N., Yukinaga H., Ebisuya M., Shiki T., Kurihara T., Kioka N., Kambe T., Nagao M., Nishida E.et al. .. The closely related RNA helicases, UAP56 and URH49, preferentially form distinct mRNA export machineries and coordinately regulate mitotic progression. Mol. Biol. Cell. 2010; 21:2953–2965. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

