Site-directed mutagenesis reveals the interplay between stability, structure, and enzymatic activity in RidA from Capra hircus
- PMID: 38801230
- PMCID: PMC11129622
- DOI: 10.1002/pro.5036
Site-directed mutagenesis reveals the interplay between stability, structure, and enzymatic activity in RidA from Capra hircus
Abstract
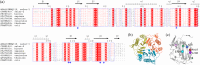
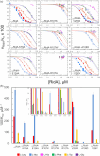
Reactive intermediate deaminase A (RidA) is a highly conserved enzyme that catalyzes the hydrolysis of 2-imino acids to the corresponding 2-keto acids and ammonia. RidA thus prevents the accumulation of such potentially harmful compounds in the cell, as exemplified by its role in the degradation of 2-aminoacrylate, formed during the metabolism of cysteine and serine, catalyzing the conversion of its stable 2-iminopyruvate tautomer into pyruvate. Capra hircus (goat) RidA (ChRidA) was the first mammalian RidA to be isolated and described. It has the typical homotrimeric fold of the Rid superfamily, characterized by remarkably high thermal stability, with three active sites located at the interface between adjacent subunits. ChRidA exhibits a broad substrate specificity with a preference for 2-iminopyruvate and other 2-imino acids derived from amino acids with non-polar non-bulky side chains. Here we report a biophysical and biochemical characterization of eight ChRidA variants obtained by site-directed mutagenesis to gain insight into the role of specific residues in protein stability and catalytic activity. Each mutant was produced in Escherichia coli cells, purified and characterized in terms of quaternary structure, thermal stability and substrate specificity. The results are rationalized in the context of the high-resolution structures obtained by x-ray crystallography.
Keywords: 2‐aminoacrylate; 2‐imino acids; RidA; metabolic damage; protein stability; reactive intermediate deaminase A; x‐ray crystallography.
© 2024 The Protein Society.
Figures


Similar articles
-
Apis mellifera RidA, a novel member of the canonical YigF/YER057c/UK114 imine deiminase superfamily of enzymes pre-empting metabolic damage.Biochem Biophys Res Commun. 2022 Aug 6;616:70-75. doi: 10.1016/j.bbrc.2022.05.062. Epub 2022 May 24. Biochem Biophys Res Commun. 2022. PMID: 35640488
-
RidA Proteins Protect against Metabolic Damage by Reactive Intermediates.Microbiol Mol Biol Rev. 2020 Jul 15;84(3):e00024-20. doi: 10.1128/MMBR.00024-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32669283 Free PMC article. Review.
-
Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage.Sci Rep. 2020 Jun 23;10(1):10135. doi: 10.1038/s41598-020-66663-w. Sci Rep. 2020. PMID: 32576850 Free PMC article.
-
The Cysteine Desulfurase IscS Is a Significant Target of 2-Aminoacrylate Damage in Pseudomonas aeruginosa.mBio. 2022 Jun 28;13(3):e0107122. doi: 10.1128/mbio.01071-22. Epub 2022 Jun 2. mBio. 2022. PMID: 35652590 Free PMC article.
-
Reactive Enamines and Imines In Vivo: Lessons from the RidA Paradigm.Trends Biochem Sci. 2019 Oct;44(10):849-860. doi: 10.1016/j.tibs.2019.04.011. Epub 2019 May 15. Trends Biochem Sci. 2019. PMID: 31103411 Free PMC article. Review.
References
-
- Asagi K, Oka T, Arao K, Suzuki I, Thakur MK, Izumi K, et al. Purification, characterization and differentiation‐dependent expression of a perchloric acid soluble protein from rat kidney. Nephron. 1998;79:80–90. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

