Mechanisms and Delivery of tRNA Therapeutics
- PMID: 38801719
- PMCID: PMC11212642
- DOI: 10.1021/acs.chemrev.4c00142
Mechanisms and Delivery of tRNA Therapeutics
Abstract
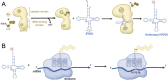
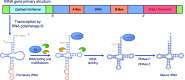
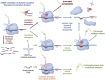
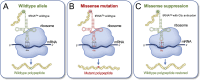
Transfer ribonucleic acid (tRNA) therapeutics will provide personalized and mutation specific medicines to treat human genetic diseases for which no cures currently exist. The tRNAs are a family of adaptor molecules that interpret the nucleic acid sequences in our genes into the amino acid sequences of proteins that dictate cell function. Humans encode more than 600 tRNA genes. Interestingly, even healthy individuals contain some mutant tRNAs that make mistakes. Missense suppressor tRNAs insert the wrong amino acid in proteins, and nonsense suppressor tRNAs read through premature stop signals to generate full length proteins. Mutations that underlie many human diseases, including neurodegenerative diseases, cancers, and diverse rare genetic disorders, result from missense or nonsense mutations. Thus, specific tRNA variants can be strategically deployed as therapeutic agents to correct genetic defects. We review the mechanisms of tRNA therapeutic activity, the nature of the therapeutic window for nonsense and missense suppression as well as wild-type tRNA supplementation. We discuss the challenges and promises of delivering tRNAs as synthetic RNAs or as gene therapies. Together, tRNA medicines will provide novel treatments for common and rare genetic diseases in humans.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

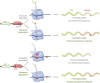
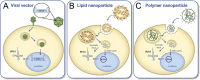
References
-
- Söll D.; Ohtsuka E.; Jones D. S.; Lohrmann R.; Hayatsu H.; Nishimura S.; Khorana H. G. Studies on polynucleotides, XLIX. Stimulation of the binding of aminoacyl-sRNA’s to ribosomes by ribotrinucleotides and a survey of codon assignments for 20 amino acids. Proc. Natl. Acad. Sci. U.S.A. 1965, 54, 1378–1385. 10.1073/pnas.54.5.1378. - DOI - PMC - PubMed
-
- Nishimura S.; Jones D. S.; Khorana H. G. Studies on polynucleotides. 48. The in vitro synthesis of a co-polypeptide containing two amino acids in alternating sequence dependent upon a DNA-like polymer containing two nucleotides in alternating sequence. J. Mol. Biol. 1965, 13, 302–324. 10.1016/S0022-2836(65)80098-5. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

