Shifts in evolutionary lability underlie independent gains and losses of root-nodule symbiosis in a single clade of plants
- PMID: 38802387
- PMCID: PMC11130336
- DOI: 10.1038/s41467-024-48036-3
Shifts in evolutionary lability underlie independent gains and losses of root-nodule symbiosis in a single clade of plants
Erratum in
-
Author Correction: Shifts in evolutionary lability underlie independent gains and losses of root-nodule symbiosis in a single clade of plants.Nat Commun. 2024 Aug 1;15(1):6491. doi: 10.1038/s41467-024-50907-8. Nat Commun. 2024. PMID: 39090137 Free PMC article. No abstract available.
Abstract
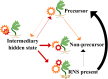
Root nodule symbiosis (RNS) is a complex trait that enables plants to access atmospheric nitrogen converted into usable forms through a mutualistic relationship with soil bacteria. Pinpointing the evolutionary origins of RNS is critical for understanding its genetic basis, but building this evolutionary context is complicated by data limitations and the intermittent presence of RNS in a single clade of ca. 30,000 species of flowering plants, i.e., the nitrogen-fixing clade (NFC). We developed the most extensive de novo phylogeny for the NFC and an RNS trait database to reconstruct the evolution of RNS. Our analysis identifies evolutionary rate heterogeneity associated with a two-step process: An ancestral precursor state transitioned to a more labile state from which RNS was rapidly gained at multiple points in the NFC. We illustrate how a two-step process could explain multiple independent gains and losses of RNS, contrary to recent hypotheses suggesting one gain and numerous losses, and suggest a broader phylogenetic and genetic scope may be required for genome-phenome mapping.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Comparative Transcriptomic Analysis of Two Actinorhizal Plants and the Legume Medicago truncatula Supports the Homology of Root Nodule Symbioses and Is Congruent With a Two-Step Process of Evolution in the Nitrogen-Fixing Clade of Angiosperms.Front Plant Sci. 2018 Oct 8;9:1256. doi: 10.3389/fpls.2018.01256. eCollection 2018. Front Plant Sci. 2018. PMID: 30349546 Free PMC article.
-
Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis.Science. 2018 Jul 13;361(6398):eaat1743. doi: 10.1126/science.aat1743. Epub 2018 May 24. Science. 2018. PMID: 29794220
-
Comparative genomics suggests that an ancestral polyploidy event leads to enhanced root nodule symbiosis in the Papilionoideae.Mol Biol Evol. 2013 Dec;30(12):2602-11. doi: 10.1093/molbev/mst152. Epub 2013 Sep 4. Mol Biol Evol. 2013. PMID: 24008584
-
Innovations in two genes kickstarted the evolution of nitrogen-fixing nodules.Curr Opin Plant Biol. 2024 Feb;77:102446. doi: 10.1016/j.pbi.2023.102446. Epub 2023 Sep 9. Curr Opin Plant Biol. 2024. PMID: 37696726 Review.
-
Evolution of NIN and NIN-like Genes in Relation to Nodule Symbiosis.Genes (Basel). 2020 Jul 11;11(7):777. doi: 10.3390/genes11070777. Genes (Basel). 2020. PMID: 32664480 Free PMC article. Review.
Cited by
-
Comparative phylogenomics and phylotranscriptomics provide insights into the genetic complexity of nitrogen-fixing root-nodule symbiosis.Plant Commun. 2024 Jan 8;5(1):100671. doi: 10.1016/j.xplc.2023.100671. Epub 2023 Aug 8. Plant Commun. 2024. PMID: 37553834 Free PMC article.
-
An integrative framework reveals widespread gene flow during the early radiation of oaks and relatives in Quercoideae (Fagaceae).J Integr Plant Biol. 2025 Apr;67(4):1119-1141. doi: 10.1111/jipb.13773. Epub 2024 Sep 19. J Integr Plant Biol. 2025. PMID: 39297574 Free PMC article.
-
Unveiling root nodulation in Tribulus terrestris and Roystonea regia via metagenomics analysis.Mol Genet Genomics. 2024 Dec 28;300(1):9. doi: 10.1007/s00438-024-02218-2. Mol Genet Genomics. 2024. PMID: 39731654
-
Dancing to a different tune, can we switch from chemical to biological nitrogen fixation for sustainable food security?PLoS Biol. 2023 Mar 14;21(3):e3001982. doi: 10.1371/journal.pbio.3001982. eCollection 2023 Mar. PLoS Biol. 2023. PMID: 36917569 Free PMC article.
-
Enacting partner specificity in legume-rhizobia symbioses.aBIOTECH. 2024 Dec 23;6(2):311-327. doi: 10.1007/s42994-024-00193-1. eCollection 2025 Jun. aBIOTECH. 2024. PMID: 40641649 Free PMC article. Review.
References
-
- Darwin, C. On the Origin of Species: By Means of Natural Selection. (Courier Corporation, 2006).
MeSH terms
LinkOut - more resources
Full Text Sources

