NG2 glia protect against prion neurotoxicity by inhibiting microglia-to-neuron prostaglandin E2 signaling
- PMID: 38802591
- PMCID: PMC11303249
- DOI: 10.1038/s41593-024-01663-x
NG2 glia protect against prion neurotoxicity by inhibiting microglia-to-neuron prostaglandin E2 signaling
Abstract
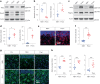
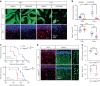
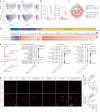
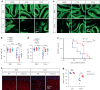
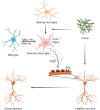
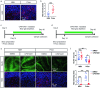
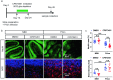
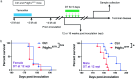
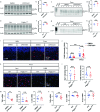
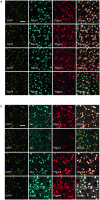
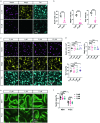
Oligodendrocyte-lineage cells, including NG2 glia, undergo prominent changes in various neurodegenerative disorders. Here, we identify a neuroprotective role for NG2 glia against prion toxicity. NG2 glia were activated after prion infection in cerebellar organotypic cultured slices (COCS) and in brains of prion-inoculated mice. In both model systems, depletion of NG2 glia exacerbated prion-induced neurodegeneration and accelerated prion pathology. Loss of NG2 glia enhanced the biosynthesis of prostaglandin E2 (PGE2) by microglia, which augmented prion neurotoxicity through binding to the EP4 receptor. Pharmacological or genetic inhibition of PGE2 biosynthesis attenuated prion-induced neurodegeneration in COCS and mice, reduced the enhanced neurodegeneration in NG2-glia-depleted COCS after prion infection, and dampened the acceleration of prion disease in NG2-glia-depleted mice. These data unveil a non-cell-autonomous interaction between NG2 glia and microglia in prion disease and suggest that PGE2 signaling may represent an actionable target against prion diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Liu, Y. & Aguzzi, A. Immunotherapy for neurodegeneration? Science364, 130–131 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- 670958/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 179040/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- FK Liu 19-042/Universität Zürich (University of Zurich)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

