MUC1-C regulates NEAT1 lncRNA expression and paraspeckle formation in cancer progression
- PMID: 38802648
- PMCID: PMC11226401
- DOI: 10.1038/s41388-024-03068-3
MUC1-C regulates NEAT1 lncRNA expression and paraspeckle formation in cancer progression
Abstract
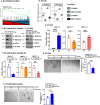
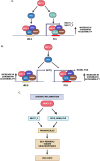
The MUC1 gene evolved in mammals for adaptation of barrier tissues in response to infections and damage. Paraspeckles are nuclear bodies formed on the NEAT1 lncRNA in response to loss of homeostasis. There is no known intersection of MUC1 with NEAT1 or paraspeckles. Here, we demonstrate that the MUC1-C subunit plays an essential role in regulating NEAT1 expression. MUC1-C activates the NEAT1 gene with induction of the NEAT1_1 and NEAT1_2 isoforms by NF-κB- and MYC-mediated mechanisms. MUC1-C/MYC signaling also induces expression of the SFPQ, NONO and FUS RNA binding proteins (RBPs) that associate with NEAT1_2 and are necessary for paraspeckle formation. MUC1-C integrates activation of NEAT1 and RBP-encoding genes by recruiting the PBAF chromatin remodeling complex and increasing chromatin accessibility of their respective regulatory regions. We further demonstrate that MUC1-C and NEAT1 form an auto-inductive pathway that drives common sets of genes conferring responses to inflammation and loss of homeostasis. Of functional significance, we find that the MUC1-C/NEAT1 pathway is of importance for the cancer stem cell (CSC) state and anti-cancer drug resistance. These findings identify a previously unrecognized role for MUC1-C in the regulation of NEAT1, RBPs, and paraspeckles that has been co-opted in promoting cancer progression.
© 2024. The Author(s).
Conflict of interest statement
DK has equity interests in Genus Oncology and is a paid consultant to CanBas. The other authors declared no competing interests.
Figures

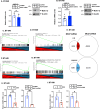
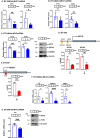
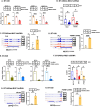

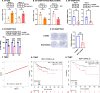
References
MeSH terms
Substances
Grants and funding
- R21 CA262991/CA/NCI NIH HHS/United States
- R01 CA097098/CA/NCI NIH HHS/United States
- U01 CA233084/CA/NCI NIH HHS/United States
- CA233084/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- CA262991/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

