LinG3D: visualizing the spatio-temporal dynamics of clonal evolution
- PMID: 38802748
- PMCID: PMC11131251
- DOI: 10.1186/s12859-024-05813-7
LinG3D: visualizing the spatio-temporal dynamics of clonal evolution
Abstract
Background: Cancers are spatially heterogenous, thus their clonal evolution, especially following anti-cancer treatments, depends on where the mutated cells are located within the tumor tissue. For example, cells exposed to different concentrations of drugs, such as cells located near the vessels in contrast to those residing far from the vasculature, can undergo a different evolutionary path. However, classical representations of cell lineage trees do not account for this spatial component of emerging cancer clones. Here, we propose routines to trace spatial and temporal clonal evolution in computer simulations of the tumor evolution models.
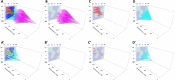
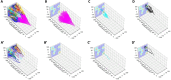
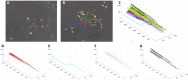
Results: The LinG3D (Lineage Graphs in 3D) is an open-source collection of routines (in MATLAB, Python, and R) that enables spatio-temporal visualization of clonal evolution in a two-dimensional tumor slice from computer simulations of the tumor evolution models. These routines draw traces of tumor clones in both time and space, and may include a projection of a selected microenvironmental factor, such as the drug or oxygen distribution within the tumor, if such a microenvironmental factor is used in the tumor evolution model. The utility of LinG3D has been demonstrated through examples of simulated tumors with different number of clones and, additionally, in experimental colony growth assay.
Conclusions: This routine package extends the classical lineage trees, that show cellular clone relationships in time, by adding the space component to show the locations of cellular clones within the 2D tumor tissue patch from computer simulations of tumor evolution models.
Keywords: 3D visualization; Lineage trees; Mathematical modeling of tumor evolution; Spatial clone distribution.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Update of
-
Visualizing the Spatio-Temporal Dynamics of Clonal Evolution with LinG3D software.bioRxiv [Preprint]. 2024 Mar 7:2024.03.05.583631. doi: 10.1101/2024.03.05.583631. bioRxiv. 2024. Update in: BMC Bioinformatics. 2024 May 27;25(1):201. doi: 10.1186/s12859-024-05813-7. PMID: 38496472 Free PMC article. Updated. Preprint.
References
-
- MATLAB. phytree routine documentation and examples. 2021 https://www.mathworks.com/help/bioinfo/ref/phytree.html
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

