CircPIAS1 promotes hepatocellular carcinoma progression by inhibiting ferroptosis via the miR-455-3p/NUPR1/FTH1 axis
- PMID: 38802795
- PMCID: PMC11131253
- DOI: 10.1186/s12943-024-02030-x
CircPIAS1 promotes hepatocellular carcinoma progression by inhibiting ferroptosis via the miR-455-3p/NUPR1/FTH1 axis
Abstract
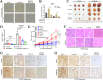
Background: The role of circRNAs in hepatocellular carcinoma (HCC) progression remains unclear. CircPIAS1 (circBase ID: hsa_circ_0007088) was identified as overexpressed in HCC cases through bioinformatics analysis. This study aimed to investigate the oncogenic properties and mechanisms of circPIAS1 in HCC development.
Methods: Functional analyses were conducted to assess circPIAS1's impact on HCC cell proliferation, migration, and ferroptosis. Xenograft mouse models were employed to evaluate circPIAS1's effects on tumor growth and pulmonary metastasis in vivo. Bioinformatics analysis, RNA immunoprecipitation, and luciferase reporter assays were utilized to elucidate the molecular pathways influenced by circPIAS1. Additional techniques, including RNA pulldown, fluorescence in situ hybridization (FISH), chromatin immunoprecipitation (ChIP), qPCR, and western blotting, were used to further explore the underlying mechanisms.
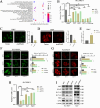
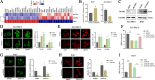
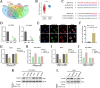
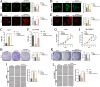
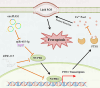
Results: CircPIAS1 expression was elevated in HCC tissues and cells. Silencing circPIAS1 suppressed HCC cell proliferation and migration both in vitro and in vivo. Mechanically, circPIAS1 overexpression inhibited ferroptosis by competitively binding to miR-455-3p, leading to upregulation of Nuclear Protein 1 (NUPR1). Furthermore, NUPR1 promoted FTH1 transcription, enhancing iron storage in HCC cells and conferring resistance to ferroptosis. Treatment with ZZW-115, an NUPR1 inhibitor, reversed the tumor-promoting effects of circPIAS1 and sensitized HCC cells to lenvatinib.
Conclusion: This study highlights the critical role of circPIAS1 in HCC progression through modulation of ferroptosis. Targeting the circPIAS1/miR-455-3p/NUPR1/FTH1 regulatory axis may represent a promising therapeutic strategy for HCC.
Keywords: Ferroptosis; Hepatocellular carcinoma; NUPR1; circPIAS1; miR-455-3p.
© 2024. The Author(s).
Conflict of interest statement
The authors have declared that no competing interest exists.
Figures



References
MeSH terms
Substances
Grants and funding
- 82070612/National Natural Science Foundation of China
- 2021A1515010306/Natural Science Foundation of Guangdong Province
- 2023B03J0154 and 2023B01J1007/Guangzhou Science and Technology Program Key Projects
- 2019B110233002/Research and Development Planned Project in Key Areas of Guangdong Province
- NA/Major Talent Training Project of the Third Affiliated Hospital of Sun Yat-sen University
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

