Impact of PEDV infection on the biological characteristics of porcine intestinal exosomes
- PMID: 38803376
- PMCID: PMC11128675
- DOI: 10.3389/fmicb.2024.1392450
Impact of PEDV infection on the biological characteristics of porcine intestinal exosomes
Abstract
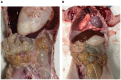
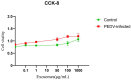
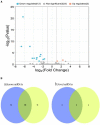
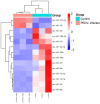
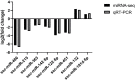
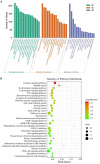
Porcine epidemic diarrhea (PED) is a highly contagious intestinal infection primarily affecting pigs. It is caused by the porcine epidemic diarrhea virus (PEDV). PEDV targets the villus tissue cells in the small intestine and mesenteric lymph nodes, resulting in shortened intestinal villi and, in extreme cases, causing necrosis of the intestinal lining. Moreover, PEDV infection can disrupt the balance of the intestinal microflora, leading to an overgrowth of harmful bacteria like Escherichia coli. Exosomes, tiny membrane vesicles ranging from 30 to 150 nm in size, contain a complex mixture of RNA and proteins. MicroRNA (miRNA) regulates various cell signaling, development, and disease progression processes. This study extracted exosomes from both groups and performed high-throughput miRNA sequencing and bioinformatics techniques to investigate differences in miRNA expression within exosomes isolated from PEDV-infected porcine small intestine tissue compared to healthy controls. Notably, two miRNA types displayed upregulation in infected exosomes, while 12 exhibited downregulation. These findings unveil abnormal miRNA regulation patterns in PEDV-infected intestinal exosomes, shedding light on the intricate interplay between PEDV and its host. This will enable further exploration of the relationship between these miRNA changes and signaling pathways, enlightening PEDV pathogenesis and potential therapeutic targets.
Keywords: exosomes; microRNA; porcine epidemic diarrhea virus; porcine intestinal tissue damage; ultracentrifugation.
Copyright © 2024 Wu, Su, Ma, Wang, Luo, EI-Ashram, Alajmi and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Characteristics of the MicroRNA Expression Profile of Exosomes Released by Vero Cells Infected with Porcine Epidemic Diarrhea Virus.Viruses. 2022 Apr 13;14(4):806. doi: 10.3390/v14040806. Viruses. 2022. PMID: 35458536 Free PMC article.
-
LncRNA446 Regulates Tight Junctions by Inhibiting the Ubiquitinated Degradation of Alix after Porcine Epidemic Diarrhea Virus Infection.J Virol. 2023 Mar 30;97(3):e0188422. doi: 10.1128/jvi.01884-22. Epub 2023 Feb 15. J Virol. 2023. PMID: 36790206 Free PMC article.
-
Porcine Intestinal Enteroids: a New Model for Studying Enteric Coronavirus Porcine Epidemic Diarrhea Virus Infection and the Host Innate Response.J Virol. 2019 Feb 19;93(5):e01682-18. doi: 10.1128/JVI.01682-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541861 Free PMC article.
-
Porcine Epidemic Diarrhea: Causative Agent, Epidemiology, Clinical Characteristics, and Treatment Strategy Targeting Main Protease.Protein Pept Lett. 2022;29(5):392-407. doi: 10.2174/0929866529666220316145149. Protein Pept Lett. 2022. PMID: 35297340 Review.
-
A Comprehensive View on the Host Factors and Viral Proteins Associated With Porcine Epidemic Diarrhea Virus Infection.Front Microbiol. 2021 Dec 7;12:762358. doi: 10.3389/fmicb.2021.762358. eCollection 2021. Front Microbiol. 2021. PMID: 34950116 Free PMC article. Review.
References
-
- Buddingh K. (2015). Exosomes in dengue virus infection. J. Extracell. Vesicles 4:e12642.
-
- Colombo M. (2014). Biogenesis, secretion, and uptake of exosomes. J. Extracell. Vesicles 2:18911.
-
- Conner S. D., Schmid S. L. (2003). The biogenesis of exosomes. Traffic 4, 118–126.
-
- Gaudin Y. (2011). HTLV-1-infected T cells release exosomes containing viral RNA and proteins. J. Virol. 85, 12738–12748.
LinkOut - more resources
Full Text Sources

