Autism risk gene Cul3 alters neuronal morphology via caspase-3 activity in mouse hippocampal neurons
- PMID: 38803442
- PMCID: PMC11129687
- DOI: 10.3389/fncel.2024.1320784
Autism risk gene Cul3 alters neuronal morphology via caspase-3 activity in mouse hippocampal neurons
Abstract
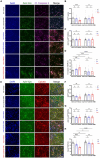
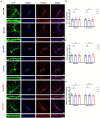
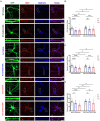
Autism Spectrum Disorders (ASDs) are neurodevelopmental disorders (NDDs) in which children display differences in social interaction/communication and repetitive stereotyped behaviors along with variable associated features. Cul3, a gene linked to ASD, encodes CUL3 (CULLIN-3), a protein that serves as a key component of a ubiquitin ligase complex with unclear function in neurons. Cul3 homozygous deletion in mice is embryonic lethal; thus, we examine the role of Cul3 deletion in early synapse development and neuronal morphology in hippocampal primary neuronal cultures. Homozygous deletion of Cul3 significantly decreased dendritic complexity and dendritic length, as well as axon formation. Synaptic spine density significantly increased, mainly in thin and stubby spines along with decreased average spine volume in Cul3 knockouts. Both heterozygous and homozygous knockout of Cul3 caused significant reductions in the density and colocalization of gephyrin/vGAT puncta, providing evidence of decreased inhibitory synapse number, while excitatory synaptic puncta vGulT1/PSD95 density remained unchanged. Based on previous studies implicating elevated caspase-3 after Cul3 deletion, we demonstrated increased caspase-3 in our neuronal cultures and decreased neuronal cell viability. We then examined the efficacy of the caspase-3 inhibitor Z-DEVD-FMK to rescue the decrease in neuronal cell viability, demonstrating reversal of the cell viability phenotype with caspase-3 inhibition. Studies have also implicated caspase-3 in neuronal morphological changes. We found that caspase-3 inhibition largely reversed the dendrite, axon, and spine morphological changes along with the inhibitory synaptic puncta changes. Overall, these data provide additional evidence that Cul3 regulates the formation or maintenance of cell morphology, GABAergic synaptic puncta, and neuronal viability in developing hippocampal neurons in culture.
Keywords: axon; caspase-3; cullin-3; dendrite; spine; synapse.
Copyright © 2024 Xia, Singh, Wang, Xuan, Singer and Powell.
Conflict of interest statement
CP has been funded in the past by Novartis and reimbursed for travel to speak once each at Dainippon Sumitomo Pharma Co., Pfizer, Roche, Astra-Zeneca, and Psychogenics. CP has also served once as a paid expert witness for attorneys representing Johnson & Johnson, Inc., among other defendants. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Effects of heterozygous deletion of autism-related gene Cullin-3 in mice.PLoS One. 2023 Jul 10;18(7):e0283299. doi: 10.1371/journal.pone.0283299. eCollection 2023. PLoS One. 2023. PMID: 37428799 Free PMC article.
-
Deficiency of Cullin 3, a Protein Encoded by a Schizophrenia and Autism Risk Gene, Impairs Behaviors by Enhancing the Excitability of Ventral Tegmental Area (VTA) DA Neurons.J Neurosci. 2023 Sep 6;43(36):6249-6267. doi: 10.1523/JNEUROSCI.0247-23.2023. Epub 2023 Aug 9. J Neurosci. 2023. PMID: 37558490 Free PMC article.
-
Autism-linked Cullin3 germline haploinsufficiency impacts cytoskeletal dynamics and cortical neurogenesis through RhoA signaling.Mol Psychiatry. 2021 Jul;26(7):3586-3613. doi: 10.1038/s41380-021-01052-x. Epub 2021 Mar 16. Mol Psychiatry. 2021. PMID: 33727673 Free PMC article.
-
Current trends of high-risk gene Cul3 in neurodevelopmental disorders.Front Psychiatry. 2023 Jul 28;14:1215110. doi: 10.3389/fpsyt.2023.1215110. eCollection 2023. Front Psychiatry. 2023. PMID: 37575562 Free PMC article. Review.
-
The Autism Protein Ube3A/E6AP Remodels Neuronal Dendritic Arborization via Caspase-Dependent Microtubule Destabilization.J Neurosci. 2018 Jan 10;38(2):363-378. doi: 10.1523/JNEUROSCI.1511-17.2017. Epub 2017 Dec 4. J Neurosci. 2018. PMID: 29175955 Free PMC article.
References
-
- Amar M., Pramod A. B., Yu N. K., Herrera V. M., Qiu L. R., Moran-Losada P., et al. . (2021). Autism-linked Cullin 3 germline haploinsufficiency impacts cytoskeletal dynamics and cortical neurogenesis through rho a signaling. Mol. Psychiatry 26, 3586–3613. doi: 10.1038/s41380-021-01052-x, PMID: - DOI - PMC - PubMed
-
- American Psychiatric Association . Task force on DSM-IV. diagnostic and statistical manual of mental disorders: DSM-IV-TR. 4th Washington, DC: American Psychiatric Association; (2000).
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

