Discovery of biomarkers in the psoriasis through machine learning and dynamic immune infiltration in three types of skin lesions
- PMID: 38803495
- PMCID: PMC11128609
- DOI: 10.3389/fimmu.2024.1388690
Discovery of biomarkers in the psoriasis through machine learning and dynamic immune infiltration in three types of skin lesions
Abstract
Introduction: Psoriasis is a chronic skin disease characterized by unique scaling plaques. However, during the acute phase, psoriatic lesions exhibit eczematous changes, making them difficult to distinguish from atopic dermatitis, which poses challenges for the selection of biological agents. This study aimed to identify potential diagnostic genes in psoriatic lesions and investigate their clinical significance.
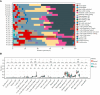
Methods: GSE182740 datasets from the GEO database were analyzed for differential analysis; machine learning algorithms (SVM-RFE and LASSO regression models) are used to screen for diagnostic markers; CIBERSORTx is used to determine the dynamic changes of 22 different immune cell components in normal skin lesions, psoriatic non-lesional skin, and psoriatic lesional skin, as well as the expression of the diagnostic genes in 10 major immune cells, and real-time quantitative polymerase chain reaction (RT-qPCR) and immunohistochemistry are used to validate results.
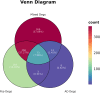
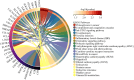
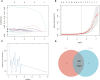
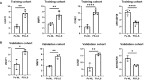
Results: We obtained 580 differentially expressed genes (DEGs) in the skin lesion and non-lesion of psoriasis patients, 813 DEGs in mixed patients between non-lesions and lesions, and 96 DEGs in the skin lesion and non-lesion of atopic dermatitis, respectively. Then 144 specific DEGs in psoriasis via a Veen diagram were identified. Ultimately, UGGT1, CCNE1, MMP9 and ARHGEF28 are identified for potential diagnostic genes from these 144 specific DEGs. The value of the selected diagnostic genes was verified by receiver operating characteristic (ROC) curves with expanded samples. The the area under the ROC curve (AUC) exceeded 0.7 for the four diagnosis genes. RT-qPCR results showed that compared to normal human epidermis, the expression of UGGT1, CCNE1, and MMP9 was significantly increased in patients with psoriasis, while ARHGEF28 expression was significantly decreased. Notably, the results of CIBERSORTx showed that CCNE1 was highly expressed in CD4+ T cells and neutrophils, ARHGEF28 was also expressed in mast cells. Additionally, CCNE1 was strongly correlated with IL-17/CXCL8/9/10 and CCL20. Immunohistochemical results showed increased nuclear expression of CCNE1 in psoriatic epidermal cells relative to normal.
Conclusion: Based on the performance of the four genes in ROC curves and their expression in immune cells from patients with psoriasis, we suggest that CCNE1 possess higher diagnostic value.
Keywords: atopic dermatitis; diagnosis genes; immune infiltration; machine learning algorithm; psoriasis.
Copyright © 2024 Zhou, Zhou, Luo and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Yu S, Li Y, Zhou Y, Follansbee T, Hwang ST. Immune mediators and therapies for pruritus in atopic dermatitis and psoriasis. J Cutaneous Immunol Allergy. (2019) 2:4–14. doi: 10.1002/cia2.12049 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

