Emerging Approaches to Profile Accessible Chromatin from Formalin-Fixed Paraffin-Embedded Sections
- PMID: 38804369
- PMCID: PMC11130958
- DOI: 10.3390/epigenomes8020020
Emerging Approaches to Profile Accessible Chromatin from Formalin-Fixed Paraffin-Embedded Sections
Abstract
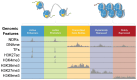
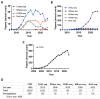
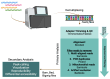
Nucleosomes are non-uniformly distributed across eukaryotic genomes, with stretches of 'open' chromatin strongly associated with transcriptionally active promoters and enhancers. Understanding chromatin accessibility patterns in normal tissue and how they are altered in pathologies can provide critical insights to development and disease. With the advent of high-throughput sequencing, a variety of strategies have been devised to identify open regions across the genome, including DNase-seq, MNase-seq, FAIRE-seq, ATAC-seq, and NicE-seq. However, the broad application of such methods to FFPE (formalin-fixed paraffin-embedded) tissues has been curtailed by the major technical challenges imposed by highly fixed and often damaged genomic material. Here, we review the most common approaches for mapping open chromatin regions, recent optimizations to overcome the challenges of working with FFPE tissue, and a brief overview of a typical data pipeline with analysis considerations.
Keywords: FFPE; chromatin; nucleosome; nucleosome-depleted region; nucleosome-free region.
Conflict of interest statement
New England Biolabs (NEB) and EpiCypher are engaged in the commercial development of NicE-seq based approaches. V.U.S.K., B.J.V., M.W.C. and M.-C.K. are employed by (and own shares in) EpiCypher. K.R., S.S., P.-O.E. and S.P. are employed by (and own shares in) NEB. M.-C.K. is a board member of EpiCypher.
Figures

Similar articles
-
Defining Regulatory Elements in the Human Genome Using Nucleosome Occupancy and Methylome Sequencing (NOMe-Seq).Methods Mol Biol. 2018;1766:209-229. doi: 10.1007/978-1-4939-7768-0_12. Methods Mol Biol. 2018. PMID: 29605855 Free PMC article.
-
Mapping Genome-wide Accessible Chromatin in Primary Human T Lymphocytes by ATAC-Seq.J Vis Exp. 2017 Nov 13;(129):56313. doi: 10.3791/56313. J Vis Exp. 2017. PMID: 29155775 Free PMC article.
-
FFPE-ATAC: A Highly Sensitive Method for Profiling Chromatin Accessibility in Formalin-Fixed Paraffin-Embedded Samples.Curr Protoc. 2022 Aug;2(8):e535. doi: 10.1002/cpz1.535. Curr Protoc. 2022. PMID: 35994571 Free PMC article.
-
[Advances in assay for transposase-accessible chromatin with high-throughput sequencing].Yi Chuan. 2020 Apr 20;42(4):333-346. doi: 10.16288/j.yczz.19-279. Yi Chuan. 2020. PMID: 32312702 Review. Chinese.
-
Chromatin accessibility: a window into the genome.Epigenetics Chromatin. 2014 Nov 20;7(1):33. doi: 10.1186/1756-8935-7-33. eCollection 2014. Epigenetics Chromatin. 2014. PMID: 25473421 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

