OrganoIDNet: a deep learning tool for identification of therapeutic effects in PDAC organoid-PBMC co-cultures from time-resolved imaging data
- PMID: 38805131
- PMCID: PMC11850476
- DOI: 10.1007/s13402-024-00958-2
OrganoIDNet: a deep learning tool for identification of therapeutic effects in PDAC organoid-PBMC co-cultures from time-resolved imaging data
Abstract
Purpose: Pancreatic Ductal Adenocarcinoma (PDAC) remains a challenging disease due to its complex biology and aggressive behavior with an urgent need for efficient therapeutic strategies. To assess therapy response, pre-clinical PDAC organoid-based models in combination with accurate real-time monitoring are required.
Methods: We established stable live-imaging organoid/peripheral blood mononuclear cells (PBMCs) co-cultures and introduced OrganoIDNet, a deep-learning-based algorithm, capable of analyzing bright-field images of murine and human patient-derived PDAC organoids acquired with live-cell imaging. We investigated the response to the chemotherapy gemcitabine in PDAC organoids and the PD-L1 inhibitor Atezolizumab, cultured with or without HLA-matched PBMCs over time. Results obtained with OrganoIDNet were validated with the endpoint proliferation assay CellTiter-Glo.
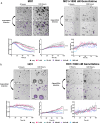
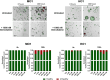
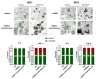
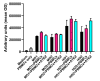
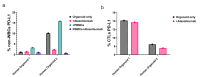
Results: Live cell imaging in combination with OrganoIDNet accurately detected size-specific drug responses of organoids to gemcitabine over time, showing that large organoids were more prone to cytotoxic effects. This approach also allowed distinguishing between healthy and unhealthy status and measuring eccentricity as organoids' reaction to therapy. Furthermore, imaging of a new organoids/PBMCs sandwich-based co-culture enabled longitudinal analysis of organoid responses to Atezolizumab, showing an increased potency of PBMCs tumor-killing in an organoid-individual manner when Atezolizumab was added.
Conclusion: Optimized PDAC organoid imaging analyzed by OrganoIDNet represents a platform capable of accurately detecting organoid responses to standard PDAC chemotherapy over time. Moreover, organoid/immune cell co-cultures allow monitoring of organoid responses to immunotherapy, offering dynamic insights into treatment behavior within a co-culture setting with PBMCs. This setup holds promise for real-time assessment of immunotherapeutic effects in individual patient-derived PDAC organoids.
Keywords: Artificial intelligence; Co-cultures; Gemcitabine; Immunotherapy; Organoids; PDAC.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures







References
-
- R.L. Siegel, K.D. Miller, A. Jemal, Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

