What shapes template-matching performance in cryogenic electron tomography in situ?
- PMID: 38805246
- PMCID: PMC11154592
- DOI: 10.1107/S2059798324004303
What shapes template-matching performance in cryogenic electron tomography in situ?
Abstract
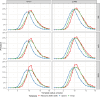
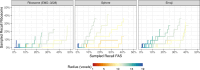
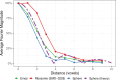
The detection of specific biological macromolecules in cryogenic electron tomography data is frequently approached by applying cross-correlation-based 3D template matching. To reduce computational cost and noise, high binning is used to aggregate voxels before template matching. This remains a prevalent practice in both practical applications and methods development. Here, the relation between template size, shape and angular sampling is systematically evaluated to identify ribosomes in a ground-truth annotated data set. It is shown that at the commonly used binning, a detailed subtomogram average, a sphere and a heart emoji result in near-identical performance. These findings indicate that with current template-matching practices macromolecules can only be detected with high precision if their shape and size are sufficiently different from the background. Using theoretical considerations, the experimental results are rationalized and it is discussed why primarily low-frequency information remains at high binning and that template matching fails to be accurate because similarly shaped and sized macromolecules have similar low-frequency spectra. These challenges are discussed and potential enhancements for future template-matching methodologies are proposed.
Keywords: computer vision; cryo-electron tomography; particle picking; template matching.
open access.
Figures


Similar articles
-
Simulating cryo electron tomograms of crowded cell cytoplasm for assessment of automated particle picking.BMC Bioinformatics. 2016 Oct 5;17(1):405. doi: 10.1186/s12859-016-1283-3. BMC Bioinformatics. 2016. PMID: 27716029 Free PMC article.
-
Locating macromolecular assemblies in cells by 2D template matching with cisTEM.Elife. 2021 Jun 11;10:e68946. doi: 10.7554/eLife.68946. Elife. 2021. PMID: 34114559 Free PMC article.
-
Deep learning improves macromolecule identification in 3D cellular cryo-electron tomograms.Nat Methods. 2021 Nov;18(11):1386-1394. doi: 10.1038/s41592-021-01275-4. Epub 2021 Oct 21. Nat Methods. 2021. PMID: 34675434
-
In Situ Cryo-Electron Tomography: A Post-Reductionist Approach to Structural Biology.J Mol Biol. 2016 Jan 29;428(2 Pt A):332-343. doi: 10.1016/j.jmb.2015.09.030. Epub 2015 Oct 9. J Mol Biol. 2016. PMID: 26456135 Review.
-
Combining high throughput and high quality for cryo-electron microscopy data collection.Acta Crystallogr D Struct Biol. 2020 Aug 1;76(Pt 8):724-728. doi: 10.1107/S2059798320008347. Epub 2020 Jul 27. Acta Crystallogr D Struct Biol. 2020. PMID: 32744254 Free PMC article. Review.
Cited by
-
PickET: An unsupervised method for localizing macromolecules in cryo-electron tomograms.bioRxiv [Preprint]. 2025 Aug 21:2025.08.20.671250. doi: 10.1101/2025.08.20.671250. bioRxiv. 2025. PMID: 40894555 Free PMC article. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

