Circular Clustering With Polar Coordinate Reconstruction
- PMID: 38805330
- PMCID: PMC11568864
- DOI: 10.1109/TCBB.2024.3406341
Circular Clustering With Polar Coordinate Reconstruction
Abstract
There is a growing interest in characterizing circular data found in biological systems. Such data are wide-ranging and varied, from the signal phase in neural recordings to nucleotide sequences in round genomes. Traditional clustering algorithms are often inadequate due to their limited ability to distinguish differences in the periodic component θ. Current clustering schemes for polar coordinate systems have limitations, such as being only angle-focused or lacking generality. To overcome these limitations, we propose a new analysis framework that utilizes projections onto a cylindrical coordinate system to represent objects in a polar coordinate system optimally. Using the mathematical properties of circular data, we show that our approach always finds the correct clustering result within the reconstructed dataset, given sufficient periodic repetitions of the data. This framework is generally applicable and adaptable to most state-of-the-art clustering algorithms. We demonstrate on synthetic and real data that our method generates more appropriate and consistent clustering results than standard methods. In summary, our proposed analysis framework overcomes the limitations of existing polar coordinate-based clustering methods and provides an accurate and efficient way to cluster circular data.
Figures
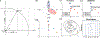
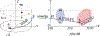





References
-
- Anand S, Padmanabham P, and Govardhan A, “Effect of distance measures on partitional clustering algorithms using transportation data,” Int. J. Comput. Sci. Inf. Technol, vol. 6, no. 6, pp. 5308–5312, 2015.
-
- Merrell R and Diaz D, “Clustering analyses methods: Strategies and algorithms,” Rev. Theor. Sci, vol. 4, no. 2, pp. 153–158, 2016.
-
- Patil YS and Joshi MR, “Clustering with polar coordinates system: Exploring possibilities,” in Smart Intelligent Computing and Applications, Berlin, Germany: Springer, 2019, pp. 553–560.
-
- Jain AK, Murty MN, and Flynn PJ, “Data clustering: A review,” ACM Comput. Surv, vol. 31, no. 3, pp. 264–323, 1999.
-
- Fell J and Axmacher N, “The role of phase synchronization in memory processes,” Nature Rev. Neurosci, vol. 12, no. 2, pp. 105–118, 2011. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

