Clock-dependent chromatin accessibility rhythms regulate circadian transcription
- PMID: 38805552
- PMCID: PMC11161047
- DOI: 10.1371/journal.pgen.1011278
Clock-dependent chromatin accessibility rhythms regulate circadian transcription
Abstract
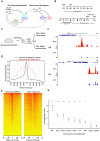
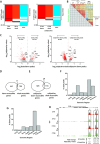
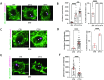
Chromatin organization plays a crucial role in gene regulation by controlling the accessibility of DNA to transcription machinery. While significant progress has been made in understanding the regulatory role of clock proteins in circadian rhythms, how chromatin organization affects circadian rhythms remains poorly understood. Here, we employed ATAC-seq (Assay for Transposase-Accessible Chromatin with Sequencing) on FAC-sorted Drosophila clock neurons to assess genome-wide chromatin accessibility at dawn and dusk over the circadian cycle. We observed significant oscillations in chromatin accessibility at promoter and enhancer regions of hundreds of genes, with enhanced accessibility either at dusk or dawn, which correlated with their peak transcriptional activity. Notably, genes with enhanced accessibility at dusk were enriched with E-box motifs, while those more accessible at dawn were enriched with VRI/PDP1-box motifs, indicating that they are regulated by the core circadian feedback loops, PER/CLK and VRI/PDP1, respectively. Further, we observed a complete loss of chromatin accessibility rhythms in per01 null mutants, with chromatin consistently accessible at both dawn and dusk, underscoring the critical role of Period protein in driving chromatin compaction during the repression phase at dawn. Together, this study demonstrates the significant role of chromatin organization in circadian regulation, revealing how the interplay between clock proteins and chromatin structure orchestrates the precise timing of biological processes throughout the day. This work further implies that variations in chromatin accessibility might play a central role in the generation of diverse circadian gene expression patterns in clock neurons.
Copyright: © 2024 Yuan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Update of
-
Clock-dependent chromatin accessibility rhythms regulate circadian transcription.bioRxiv [Preprint]. 2023 Aug 16:2023.08.15.553315. doi: 10.1101/2023.08.15.553315. bioRxiv. 2023. Update in: PLoS Genet. 2024 May 28;20(5):e1011278. doi: 10.1371/journal.pgen.1011278. PMID: 37645872 Free PMC article. Updated. Preprint.
References
-
- Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, et al. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell. 2018;173(6):1535–48 e16. Epub 20180426. doi: 10.1016/j.cell.2018.03.074 ; PubMed Central PMCID: PMC5989727. - DOI - PMC - PubMed
-
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13(8):903–13. Epub 20110724. doi: 10.1038/ncb2285 ; PubMed Central PMCID: PMC3155811. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

