DUVEL: an active-learning annotated biomedical corpus for the recognition of oligogenic combinations
- PMID: 38805753
- PMCID: PMC11131422
- DOI: 10.1093/database/baae039
DUVEL: an active-learning annotated biomedical corpus for the recognition of oligogenic combinations
Abstract
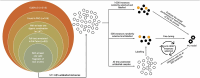
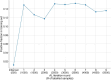
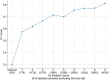
While biomedical relation extraction (bioRE) datasets have been instrumental in the development of methods to support biocuration of single variants from texts, no datasets are currently available for the extraction of digenic or even oligogenic variant relations, despite the reports in literature that epistatic effects between combinations of variants in different loci (or genes) are important to understand disease etiologies. This work presents the creation of a unique dataset of oligogenic variant combinations, geared to train tools to help in the curation of scientific literature. To overcome the hurdles associated with the number of unlabelled instances and the cost of expertise, active learning (AL) was used to optimize the annotation, thus getting assistance in finding the most informative subset of samples to label. By pre-annotating 85 full-text articles containing the relevant relations from the Oligogenic Diseases Database (OLIDA) with PubTator, text fragments featuring potential digenic variant combinations, i.e. gene-variant-gene-variant, were extracted. The resulting fragments of texts were annotated with ALAMBIC, an AL-based annotation platform. The resulting dataset, called DUVEL, is used to fine-tune four state-of-the-art biomedical language models: BiomedBERT, BiomedBERT-large, BioLinkBERT and BioM-BERT. More than 500 000 text fragments were considered for annotation, finally resulting in a dataset with 8442 fragments, 794 of them being positive instances, covering 95% of the original annotated articles. When applied to gene-variant pair detection, BiomedBERT-large achieves the highest F1 score (0.84) after fine-tuning, demonstrating significant improvement compared to the non-fine-tuned model, underlining the relevance of the DUVEL dataset. This study shows how AL may play an important role in the creation of bioRE dataset relevant for biomedical curation applications. DUVEL provides a unique biomedical corpus focusing on 4-ary relations between two genes and two variants. It is made freely available for research on GitHub and Hugging Face. Database URL: https://huggingface.co/datasets/cnachteg/duvel or https://doi.org/10.57967/hf/1571.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Bunescu R., Ge R., Kate R.J. et al. (2005) Comparative experiments on learning information extractors for proteins and their interactions. Artif. Intell. Med., 33, 139–155. - PubMed
-
- Herrero-Zazo M., Segura-Bedmar I., Martínez P. et al. (2013) The DDI corpus: an annotated corpus with pharmacological substances and drug-drug interactions. J. Biomed. Inform., 46, 914–920. - PubMed
-
- Tiktinsky A., Viswanathan V., Niezni D. et al. (2022) A dataset for N-ary relation extraction of drug combinations. In: Proceedings of the 2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, Stroudsburg, PA, USA.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

