Genomic and morphological characterization of Knufia obscura isolated from the Mars 2020 spacecraft assembly facility
- PMID: 38806503
- PMCID: PMC11133487
- DOI: 10.1038/s41598-024-61115-1
Genomic and morphological characterization of Knufia obscura isolated from the Mars 2020 spacecraft assembly facility
Abstract
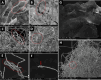
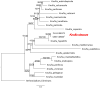
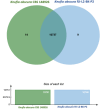
Members of the family Trichomeriaceae, belonging to the Chaetothyriales order and the Ascomycota phylum, are known for their capability to inhabit hostile environments characterized by extreme temperatures, oligotrophic conditions, drought, or presence of toxic compounds. The genus Knufia encompasses many polyextremophilic species. In this report, the genomic and morphological features of the strain FJI-L2-BK-P2 presented, which was isolated from the Mars 2020 mission spacecraft assembly facility located at the Jet Propulsion Laboratory in Pasadena, California. The identification is based on sequence alignment for marker genes, multi-locus sequence analysis, and whole genome sequence phylogeny. The morphological features were studied using a diverse range of microscopic techniques (bright field, phase contrast, differential interference contrast and scanning electron microscopy). The phylogenetic marker genes of the strain FJI-L2-BK-P2 exhibited highest similarities with type strain of Knufia obscura (CBS 148926T) that was isolated from the gas tank of a car in Italy. To validate the species identity, whole genomes of both strains (FJI-L2-BK-P2 and CBS 148926T) were sequenced, annotated, and strain FJI-L2-BK-P2 was confirmed as K. obscura. The morphological analysis and description of the genomic characteristics of K. obscura FJI-L2-BK-P2 may contribute to refining the taxonomy of Knufia species. Key morphological features are reported in this K. obscura strain, resembling microsclerotia and chlamydospore-like propagules. These features known to be characteristic features in black fungi which could potentially facilitate their adaptation to harsh environments.
Keywords: Chaetothyriales; Extremophile; Trichomeriaceae; Black fungi; Genomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

