Integration of single-cell sequencing and bulk RNA-seq to identify and develop a prognostic signature related to colorectal cancer stem cells
- PMID: 38806611
- PMCID: PMC11133358
- DOI: 10.1038/s41598-024-62913-3
Integration of single-cell sequencing and bulk RNA-seq to identify and develop a prognostic signature related to colorectal cancer stem cells
Abstract
The prognosis for patients with colorectal cancer (CRC) remains worse than expected due to metastasis, recurrence, and resistance to chemotherapy. Colorectal cancer stem cells (CRCSCs) play a vital role in tumor metastasis, recurrence, and chemotherapy resistance. However, there are currently no prognostic markers based on CRCSCs-related genes available for clinical use. In this study, single-cell transcriptome sequencing was employed to distinguish cancer stem cells (CSCs) in the CRC microenvironment and analyze their properties at the single-cell level. Subsequently, data from TCGA and GEO databases were utilized to develop a prognostic risk model for CRCSCs-related genes and validate its diagnostic performance. Additionally, functional enrichment, immune response, and chemotherapeutic drug sensitivity of the relevant genes in the risk model were investigated. Lastly, the key gene RPS17 in the risk model was identified as a potential prognostic marker and therapeutic target for further comprehensive studies. Our findings provide new insights into the prognostic treatment of CRC and offer novel perspectives for a systematic and comprehensive understanding of CRC development.
Keywords: Colorectal cancer; Colorectal cancer stem cell; Prognostic signature; RPS17; Single-cell transcriptome sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



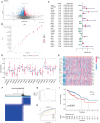

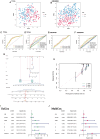


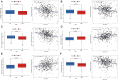
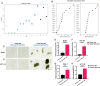

Similar articles
-
Integrative Single-Cell and Bulk RNA Sequencing Identifies a Macrophage-Related Prognostic Signature for Predicting Prognosis and Therapy Responses in Colorectal Cancer.Int J Mol Sci. 2025 Jan 19;26(2):811. doi: 10.3390/ijms26020811. Int J Mol Sci. 2025. PMID: 39859524 Free PMC article.
-
Integrated single-cell and bulk RNA-seq analysis identifies a prognostic T-cell signature in colorectal cancer.Sci Rep. 2024 Aug 30;14(1):20177. doi: 10.1038/s41598-024-70422-6. Sci Rep. 2024. PMID: 39215032 Free PMC article.
-
Integrated analysis of single-cell and bulk RNA-sequencing to predict prognosis and therapeutic response for colorectal cancer.Sci Rep. 2025 Mar 7;15(1):7986. doi: 10.1038/s41598-025-91761-y. Sci Rep. 2025. PMID: 40055390 Free PMC article.
-
Regulatory mechanisms of microRNAs in colorectal cancer and colorectal cancer stem cells.J Cell Physiol. 2020 Feb;235(2):776-789. doi: 10.1002/jcp.29042. Epub 2019 Jul 1. J Cell Physiol. 2020. PMID: 31264216 Review.
-
Prognostic gene expression profile of colorectal cancer.Gene. 2025 Jul 5;955:149433. doi: 10.1016/j.gene.2025.149433. Epub 2025 Mar 22. Gene. 2025. PMID: 40122415 Review.
Cited by
-
Redefining the tumor microenvironment with emerging therapeutic strategies.Oncol Res. 2024 Oct 16;32(11):1701-1708. doi: 10.32604/or.2024.055161. eCollection 2024. Oncol Res. 2024. PMID: 39449800 Free PMC article. Review.
-
Exploring the molecular mechanisms of macrophages in islet transplantation using single-cell analysis.Front Immunol. 2024 Aug 29;15:1407118. doi: 10.3389/fimmu.2024.1407118. eCollection 2024. Front Immunol. 2024. PMID: 39267737 Free PMC article.
-
The distinctive signature of regulatory CD4 T cells committed in the human thymus.Front Immunol. 2025 Mar 26;16:1553554. doi: 10.3389/fimmu.2025.1553554. eCollection 2025. Front Immunol. 2025. PMID: 40207234 Free PMC article.
-
Development of a prognostic risk model for colorectal cancer and association of the prognostic model with cancer stem cell and immune cell infiltration.J Gastrointest Oncol. 2025 Feb 28;16(1):77-91. doi: 10.21037/jgo-2024-985. Epub 2025 Feb 26. J Gastrointest Oncol. 2025. PMID: 40115909 Free PMC article.
-
A novel gene signature for predicting outcome in colorectal cancer patients based on tumor cell-endothelial cell interaction via single-cell sequencing and machine learning.Heliyon. 2025 Jan 24;11(3):e42237. doi: 10.1016/j.heliyon.2025.e42237. eCollection 2025 Feb 15. Heliyon. 2025. PMID: 39944338 Free PMC article.
References
-
- Patel SG, Karlitz JJ, Yen T, Lieu CH, Boland CR. The rising tide of early-onset colorectal cancer: A comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol. Hepatol. 2022;7:262–274. doi: 10.1016/S2468-1253(21)00426-X. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

