A systematic exploration of bacterial form I rubisco maximal carboxylation rates
- PMID: 38806660
- PMCID: PMC11251275
- DOI: 10.1038/s44318-024-00119-z
A systematic exploration of bacterial form I rubisco maximal carboxylation rates
Abstract
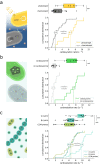
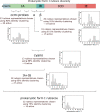
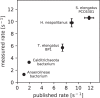
Autotrophy is the basis for complex life on Earth. Central to this process is rubisco-the enzyme that catalyzes almost all carbon fixation on the planet. Yet, with only a small fraction of rubisco diversity kinetically characterized so far, the underlying biological factors driving the evolution of fast rubiscos in nature remain unclear. We conducted a high-throughput kinetic characterization of over 100 bacterial form I rubiscos, the most ubiquitous group of rubisco sequences in nature, to uncover the determinants of rubisco's carboxylation velocity. We show that the presence of a carboxysome CO2 concentrating mechanism correlates with faster rubiscos with a median fivefold higher rate. In contrast to prior studies, we find that rubiscos originating from α-cyanobacteria exhibit the highest carboxylation rates among form I enzymes (≈10 s-1 median versus <7 s-1 in other groups). Our study systematically reveals biological and environmental properties associated with kinetic variation across rubiscos from nature.
Keywords: Carbon Fixation; Carboxysome; Cyanobacteria; Photosynthesis; Rubisco.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can J Bot. 1998;76:1052–1071.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

