Noninvasive assessment of organ-specific and shared pathways in multi-organ fibrosis using T1 mapping
- PMID: 38806679
- PMCID: PMC12574816
- DOI: 10.1038/s41591-024-03010-w
Noninvasive assessment of organ-specific and shared pathways in multi-organ fibrosis using T1 mapping
Abstract
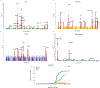
Fibrotic diseases affect multiple organs and are associated with morbidity and mortality. To examine organ-specific and shared biologic mechanisms that underlie fibrosis in different organs, we developed machine learning models to quantify T1 time, a marker of interstitial fibrosis, in the liver, pancreas, heart and kidney among 43,881 UK Biobank participants who underwent magnetic resonance imaging. In phenome-wide association analyses, we demonstrate the association of increased organ-specific T1 time, reflecting increased interstitial fibrosis, with prevalent diseases across multiple organ systems. In genome-wide association analyses, we identified 27, 18, 11 and 10 independent genetic loci associated with liver, pancreas, myocardial and renal cortex T1 time, respectively. There was a modest genetic correlation between the examined organs. Several loci overlapped across the examined organs implicating genes involved in a myriad of biologic pathways including metal ion transport (SLC39A8, HFE and TMPRSS6), glucose metabolism (PCK2), blood group antigens (ABO and FUT2), immune function (BANK1 and PPP3CA), inflammation (NFKB1) and mitosis (CENPE). Finally, we found that an increasing number of organs with T1 time falling in the top quintile was associated with increased mortality in the population. Individuals with a high burden of fibrosis in ≥3 organs had a 3-fold increase in mortality compared to those with a low burden of fibrosis across all examined organs in multivariable-adjusted analysis (hazard ratio = 3.31, 95% confidence interval 1.77-6.19; P = 1.78 × 10-4). By leveraging machine learning to quantify T1 time across multiple organs at scale, we uncovered new organ-specific and shared biologic pathways underlying fibrosis that may provide therapeutic targets.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
M.D.R.K., P.D.A, S.F.F. and P.B. are supported by grants from Bayer AG and IBM applying machine learning in cardiovascular disease. P.B. has consulted for Novartis and Prometheus Biosciences. P.B. is now employed by Flagship Pioneering. P.D.A. is now employed by Google. S.A.L. is now employed by Novartis Institutes for Biomedical Research. S.A.L. received sponsored research support from Bristol Myers Squibb / Pfizer, Bayer AG, Boehringer Ingelheim, Fitbit, and IBM, and has consulted for Bristol Myers Squibb / Pfizer, Bayer AG, Blackstone Life Sciences, and Invitae previously. P.T.E. receives sponsored research support from Bayer AG, IBM Research, Bristol Myers Squibb, Pfizer and Novo Nordisk; he has also served on advisory boards or consulted for MyoKardia and Bayer AG. L.-C.W. receives sponsored research support from IBM to the Broad Institute. The remaining authors have no disclosures.
Figures
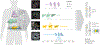


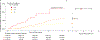
References
-
- Chen Y et al. Aging Reprograms the Hematopoietic-Vascular Niche to Impede Regeneration and Promote Fibrosis. Cell Metab 33, 395–410.e4 (2021). - PubMed
Methods Only References
-
- Li CH & Lee CK Minimum cross entropy thresholding. Pattern Recognition 26, 617–625 (1993).
MeSH terms
Substances
Grants and funding
- K08 HL159346/HL/NHLBI NIH HHS/United States
- R01 HL139731/HL/NHLBI NIH HHS/United States
- R01 HL092577/HL/NHLBI NIH HHS/United States
- 1RO1HL092577/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- T32 HL007208/HL/NHLBI NIH HHS/United States
- 1R01HL157635/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- 18SFRN34250007/AHA/American Heart Association-American Stroke Association/United States
- 5R01HL139731/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- T32HL007604/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- R01 HL157635/HL/NHLBI NIH HHS/United States
- T32 HL007604/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

