Development and verification of a manganese metabolism- and immune-related genes signature for prediction of prognosis and immune landscape in gastric cancer
- PMID: 38807601
- PMCID: PMC11131102
- DOI: 10.3389/fimmu.2024.1377472
Development and verification of a manganese metabolism- and immune-related genes signature for prediction of prognosis and immune landscape in gastric cancer
Abstract
Background: Gastric cancer (GC) poses a global health challenge due to its widespread prevalence and unfavorable prognosis. Although immunotherapy has shown promise in clinical settings, its efficacy remains limited to a minority of GC patients. Manganese, recognized for its role in the body's anti-tumor immune response, has the potential to enhance the effectiveness of tumor treatment when combined with immune checkpoint inhibitors.
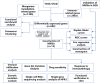
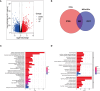
Methods: Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases was utilized to obtain transcriptome information and clinical data for GC. Unsupervised clustering was employed to stratify samples into distinct subtypes. Manganese metabolism- and immune-related genes (MIRGs) were identified in GC by univariate Cox regression and least absolute shrinkage and selection operator (LASSO) regression analysis. We conducted gene set variation analysis, and assessed the immune landscape, drug sensitivity, immunotherapy efficacy, and somatic mutations. The underlying role of NPR3 in GC was further analyzed in the single-cell RNA sequencing data and cellular experiments.
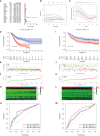
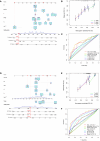
Results: GC patients were classified into four subtypes characterized by significantly different prognoses and tumor microenvironments. Thirteen genes were identified and established as MIRGs, demonstrating exceptional predictive effectiveness in GC patients. Distinct enrichment patterns of molecular functions and pathways were observed among various risk subgroups. Immune infiltration analysis revealed a significantly greater abundance of macrophages and monocytes in the high-risk group. Drug sensitivity analysis identified effective drugs for patients, while patients in the low-risk group could potentially benefit from immunotherapy. NPR3 expression was significantly downregulated in GC tissues. Single-cell RNA sequencing analysis indicated that the expression of NPR3 was distributed in endothelial cells. Cellular experiments demonstrated that NPR3 facilitated the proliferation of GC cells.
Conclusion: This is the first study to utilize manganese metabolism- and immune-related genes to identify the prognostic MIRGs for GC. The MIRGs not only reliably predicted the clinical outcome of GC patients but also hold the potential to guide future immunotherapy interventions for these patients.
Keywords: gastric cancer; immune; immunotherapy; manganese metabolism; prognostic model.
Copyright © 2024 Han, Leng, Zhao, Wang, Chen, Wang, Zhang, Li, Lu, Wang and Qi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

