COSGAP: COntainerized Statistical Genetics Analysis Pipelines
- PMID: 38808072
- PMCID: PMC11132817
- DOI: 10.1093/bioadv/vbae067
COSGAP: COntainerized Statistical Genetics Analysis Pipelines
Abstract
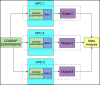
Summary: The collection and analysis of sensitive data in large-scale consortia for statistical genetics is hampered by multiple challenges, due to their non-shareable nature. Time-consuming issues in installing software frequently arise due to different operating systems, software dependencies, and limited internet access. For federated analysis across sites, it can be challenging to resolve different problems, including format requirements, data wrangling, setting up analysis on high-performance computing (HPC) facilities, etc. Easier, more standardized, automated protocols and pipelines can be solutions to overcome these issues. We have developed one such solution for statistical genetic data analysis using software container technologies. This solution, named COSGAP: "COntainerized Statistical Genetics Analysis Pipelines," consists of already established software tools placed into Singularity containers, alongside corresponding code and instructions on how to perform statistical genetic analyses, such as genome-wide association studies, polygenic scoring, LD score regression, Gaussian Mixture Models, and gene-set analysis. Using provided helper scripts written in Python, users can obtain auto-generated scripts to conduct the desired analysis either on HPC facilities or on a personal computer. COSGAP is actively being applied by users from different countries and projects to conduct genetic data analyses without spending much effort on software installation, converting data formats, and other technical requirements.
Availability and implementation: COSGAP is freely available on GitHub (https://github.com/comorment/containers) under the GPLv3 license.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
Dr. Andreassen has received speaker fees from Lundbeck, Janssen, Otsuka, and Sunovion and is a consultant to Cortechs.ai. and Precision Health. Dr. Frei is a consultant to Precision Health.
Figures

References
-
- Alles GR, Carissimi A, Schnorr LM. Assessing the computation and communication overhead of Linux containers for HPC applications. In: 2018 Symposium on High Performance Computing Systems (WSCAD), pp. 116–23. IEEE, 2018.
-
- Corfield EC, Shadrid, AA, Frei O et al. The Norwegian Mother, Father, and Child cohort study (MoBa) genotyping data resource: MoBaPsychGen pipeline v.4. bioRxiv https://www.biorxiv.org/content/10.1101/2022.06.23.496289v4,2024, preprint: not peer reviewed. - DOI
-
- Dagasso G, Yan Y, Wang L et al. Comprehensive-GWAS: a pipeline for genome-wide association studies utilizing cross-validation to assess the predictivity of genetic variations. In: 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM).2020, 1361–7.
-
- Frei O, Jangmo A, Hagen E. et al. comorment/containers: Comorment-Containers-v1.8.1 (v1.8.1). Zenodo, 2024. 10.5281/zenodo.10782180. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

