CRISPRi functional genomics in bacteria and its application to medical and industrial research
- PMID: 38809084
- PMCID: PMC11332340
- DOI: 10.1128/mmbr.00170-22
CRISPRi functional genomics in bacteria and its application to medical and industrial research
Abstract
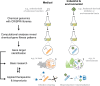
SUMMARYFunctional genomics is the use of systematic gene perturbation approaches to determine the contributions of genes under conditions of interest. Although functional genomic strategies have been used in bacteria for decades, recent studies have taken advantage of CRISPR (clustered regularly interspaced short palindromic repeats) technologies, such as CRISPRi (CRISPR interference), that are capable of precisely modulating expression of all genes in the genome. Here, we discuss and review the use of CRISPRi and related technologies for bacterial functional genomics. We discuss the strengths and weaknesses of CRISPRi as well as design considerations for CRISPRi genetic screens. We also review examples of how CRISPRi screens have defined relevant genetic targets for medical and industrial applications. Finally, we outline a few of the many possible directions that could be pursued using CRISPR-based functional genomics in bacteria. Our view is that the most exciting screens and discoveries are yet to come.
Keywords: CRISPR-Cas9; ESKAPE pathogens; antibiotics; biofuels; bioproducts; genetic screens.
Conflict of interest statement
Jason M. Peters has filed for patents related to Mobile-CRISPRi technology and bacterial promoters.
Figures



References
-
- Bosch B, DeJesus MA, Poulton NC, Zhang W, Engelhart CA, Zaveri A, Lavalette S, Ruecker N, Trujillo C, Wallach JB, Li S, Ehrt S, Chait BT, Schnappinger D, Rock JM. 2021. Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis. Cell 184:4579–4592. doi: 10.1016/j.cell.2021.06.033 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

