Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: insights from a tertiary hospital in Southern Thailand
- PMID: 38809095
- PMCID: PMC11218496
- DOI: 10.1128/spectrum.00213-24
Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: insights from a tertiary hospital in Southern Thailand
Abstract
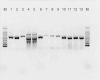
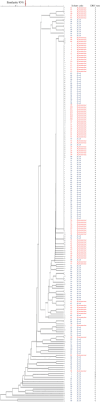
Broad-spectrum ampicillin-resistant and third-generation cephalosporin-resistant Enterobacteriaceae, particularly Escherichia coli and Klebsiella pneumoniae that have pathological features in humans, have become a global concern. This study aimed to investigate the prevalence, antimicrobial susceptibility, and molecular genetic features of extended-spectrum beta-lactamase (ESBL)-producing E. coli and K. pneumoniae isolates in Southern Thailand. Between January and August 2021, samples (n = 199) were collected from a tertiary care hospital in Southern Thailand. ESBL and AmpC-lactamase genes were identified using multiplex polymerase chain reaction (PCR). The genetic relationship between ESBL-producing E. coli and K. pneumoniae was determined using the enterobacterial repetitive intergenic consensus (ERIC) polymerase chain reaction. ESBL-producing E. coli and K. pneumoniae isolates were mostly collected from catheter urine samples of infected female patients. The ESBL production prevalence was highest in the medical wards (n = 75, 37.7%), followed by that in surgical wards (n = 64, 32.2%) and operating rooms (n = 19, 9.5%). Antimicrobial susceptibility analysis revealed that all isolates were resistant to ampicillin, cefotaxime, ceftazidime, ceftriaxone, and cefuroxime; 79.4% were resistant to ciprofloxacin; and 64.3% were resistant to trimethoprim-sulfamethoxazole. In ESBL-producing K. pneumoniae and E. coli, blaTEM (n = 57, 72.2%) and blaCTX-M (n = 61, 50.8%) genes were prominent; however, no blaVEB, blaGES, or blaPER were found in any of these isolates. Furthermore, only ESBL-producing K. pneumoniae had co-harbored blaTEM and blaSHV genes at 11.6%. The ERIC-PCR pattern of multidrug-resistant ESBL-producing strains demonstrated that the isolates were clonally related (95%). Notably, the presence of multidrug-resistant and extremely resistant ESBL producers was 83.4% and 16.6%, respectively. This study highlights the presence of blaTEM, blaCTX-M, and co-harbored genes in ESBL-producing bacterial isolates from hospitalized patients, which are associated with considerable resistance to beta-lactamase and third-generation cephalosporins.
Importance: We advocate for evidence-based guidelines and antimicrobial stewardship programs to encourage rational and appropriate antibiotic use, ultimately reducing the selection pressure for drug-resistant bacteria and lowering the likelihood of ESBL-producing bacterial infections.
Keywords: antimicrobial susceptibility analysis; enterobacterial repetitive intergenic consensus polymerase chain reaction; genetic relationship; hospitalized patients; multidrug-resistant isolates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
High prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates: A 5-year retrospective study at a Tertiary Hospital in Northern Thailand.Front Cell Infect Microbiol. 2022 Aug 8;12:955774. doi: 10.3389/fcimb.2022.955774. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36004324 Free PMC article.
-
Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae causing intra-abdominal infections from 9 tertiary hospitals in China.Diagn Microbiol Infect Dis. 2017 Jan;87(1):45-48. doi: 10.1016/j.diagmicrobio.2016.10.007. Epub 2016 Oct 7. Diagn Microbiol Infect Dis. 2017. PMID: 27773544
-
High prevalence of multidrug resistant and extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from urinary tract infections in the West region, Cameroon.BMC Infect Dis. 2025 Jan 24;25(1):115. doi: 10.1186/s12879-025-10483-8. BMC Infect Dis. 2025. PMID: 39856593 Free PMC article.
-
Diversity, functional classification and genotyping of SHV β-lactamases in Klebsiella pneumoniae.Microb Genom. 2024 Oct;10(10):001294. doi: 10.1099/mgen.0.001294. Microb Genom. 2024. PMID: 39432416 Free PMC article.
-
Emerging threats: Antimicrobial resistance in extended-spectrum beta-lactamase and carbapenem-resistant Escherichia coli.Microb Pathog. 2025 Mar;200:107275. doi: 10.1016/j.micpath.2024.107275. Epub 2025 Jan 9. Microb Pathog. 2025. PMID: 39798725 Review.
Cited by
-
Drug susceptibility of uropathogens isolated from patients treated at the Mazovian Specialized Hospital in Radom.Acta Biochim Pol. 2025 Feb 27;72:14082. doi: 10.3389/abp.2025.14082. eCollection 2025. Acta Biochim Pol. 2025. PMID: 40083641 Free PMC article.
-
Phenotypic and Genotypic Profiles of Extended-Spectrum Beta-Lactamase-Producing Multidrug-Resistant Klebsiella pneumoniae in Northeastern Thailand.Antibiotics (Basel). 2024 Sep 25;13(10):917. doi: 10.3390/antibiotics13100917. Antibiotics (Basel). 2024. PMID: 39452184 Free PMC article.
References
-
- Jenkins C, Rentenaar CRJ, Landraud L, Brisse S. 2017. 180 - Enterobacteriaceae, p 1565–1578. In Cohen JPowderly WG, Opal SM (ed), Infectious diseases, 4th ed. Elsevier, Amsterdam.
-
- Afzal A. 2017. Antibiotic resistant pattern of E. coli and Klebsiella species in Pakistan: brief overview. J Microb Biochem Tech 9:6. doi:10.4172/1948-5948.1000377 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

