Replication Stress in Activated Human NK Cells Induces Sensitivity to Apoptosis
- PMID: 38809096
- PMCID: PMC11824913
- DOI: 10.4049/jimmunol.2300843
Replication Stress in Activated Human NK Cells Induces Sensitivity to Apoptosis
Abstract
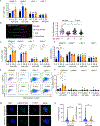
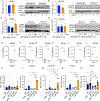
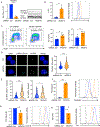
NK cells are innate immune effectors that kill virally infected or malignant cells. NK cell deficiency (NKD) occurs when NK cell development or function is impaired and variants in MCM4, GINS1, MCM10, and GINS4 result in NKD. Although NK cells are strongly impacted by mutational deficiencies in helicase proteins, the mechanisms underlying this specific susceptibility are poorly understood. In this study, we induced replication stress in activated NK cells or T cells by chemical and genetic methods. We found that the CD56bright subset of NK cells accumulates more DNA damage and replication stress during activation than do CD56dim NK cells or T cells. Aphidicolin treatment increases apoptosis of CD56bright NK cells through increased pan-caspase expression and decreases perforin expression in surviving cells. These findings show that sensitivity to replication stress affects NK cell survival and function and contributes to NKD.
Copyright © 2024 by The American Association of Immunologists, Inc.
Conflict of interest statement
Conflict-of-interest disclosure
The authors have no competing financial interests to declare.
Figures



References
-
- Cooper MA, Fehniger TA, and Caligiuri MA. 2001. The biology of human natural killer-cell subsets. Trends Immunol 22: 633–640. - PubMed
-
- Mujal AM, Delconte RB, and Sun JC. 2021. Natural Killer Cells: From Innate to Adaptive Features. Annu Rev Immunol 39: 417–447. - PubMed
-
- Cerwenka A, and Lanier LL. 2016. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol 16: 112–123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

