Epitope mapping and establishment of a blocking ELISA for mAb targeting the p72 protein of African swine fever virus
- PMID: 38809284
- PMCID: PMC11136834
- DOI: 10.1007/s00253-024-13146-x
Epitope mapping and establishment of a blocking ELISA for mAb targeting the p72 protein of African swine fever virus
Abstract
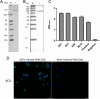
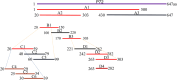
The African swine fever virus (ASFV) has the ability to infect pigs and cause a highly contagious acute fever that can result in a mortality rate as high as 100%. Due to the viral epidemic, the pig industry worldwide has suffered significant financial setbacks. The absence of a proven vaccine for ASFV necessitates the development of a sensitive and reliable serological diagnostic method, enabling laboratories to effectively and expeditiously detect ASFV infection. In this study, four strains of monoclonal antibodies (mAbs) against p72, namely, 5A1, 4C4, 8A9, and 5E10, were generated through recombinant expression of p72, the main capsid protein of ASFV, and immunized mice with it. Epitope localization was performed by truncated overlapping polypeptides. The results indicate that 5A1 and 4C4 recognized the amino acid 20-39 aa, 8A9 and 5E10 are recognized at 263-282 aa, which is consistent with the reported 265-280 aa epitopes. Conserved analysis revealed 20-39 aa is a high conservation of the epitopes in the ASFV genotypes. Moreover, a blocking ELISA assay for detection ASFV antibody based on 4C4 monoclonal antibody was developed and assessed. The receiver-operating characteristic (ROC) was performed to identify the best threshold value using 87 negative and 67 positive samples. The established test exhibited an area under the curve (AUC) of 0.9997, with a 95% confidence interval ranging from 99.87 to 100%. Furthermore, the test achieved a diagnostic sensitivity of 100% (with a 95% confidence interval of 95.72 to 100%) and a specificity of 98.51% (with a 95% confidence interval of 92.02 to 99.92%) when the threshold was set at 41.97%. The inter- and intra-batch coefficient of variation were below 10%, demonstrating the exceptional repeatability of the method. This method can detect the positive standard serum at a dilution as high as 1:512. Subsequently, an exceptional blocking ELISA assay was established with high diagnostic sensitivity and specificity, providing a novel tool for detecting ASFV antibodies. KEY POINTS: • Four strains of ASFV monoclonal antibodies against p72 were prepared and their epitopes were identified. • Blocking ELISA method was established based on monoclonal antibody 4C4 with an identified conservative epitope. • The established blocking ELISA method has a good effect on the detection of ASFV antibody.
Keywords: African swine fever virus; ELISA; Epitope mapping; Monoclonal antibodies; P72.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Bergeron HC, Glas PS, Schumann KR (2017) Diagnostic specificity of the African swine fever virus antibody detection enzyme-linked immunosorbent assay in feral and domestic pigs in the United States. Transbound Emerg Dis 64:1665–1668. 10.1111/tbed.12717 - PubMed
-
- Borca MV, Irusta P, Carrillo C, Afonso CL, Burrage T, Rock DL (1994) African swine fever virus structural protein p72 contains a conformational neutralizing epitope. Virology 201:413–418. 10.1006/viro.1994.1311 - PubMed
-
- Brookes SM, Dixon LK, Parkhouse RM (1996) Assembly of African swine fever virus: quantitative ultrastructural analysis in vitro and in vivo. Virology 224:84–92. 10.1006/viro.1996.0509 - PubMed
-
- Cadenas-Fernández E, Sánchez-Vizcaíno JM, van den Born E, Kosowska A, van Kilsdonk E, Fernández-Pacheco P, Gallardo C, Arias M, Barasona JA (2021) High doses of inactivated African swine fever virus are safe, but do not confer protection against a virulent challenge. Vaccines (basel) 9:242. 10.3390/vaccines9030242 - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2021YFD1801300/the National Key R&D Program of China
- 2021YFD1800100/the National Key R&D Program of China
- 22ZD6NA001/major science and technology projects of Gansu Province
- lzujbky-2022-ct02/Special funds for basic scientific research expenses in central colleges and universities
- 2023SDZG02/the Fundamental Research Funds for the Central Universities, the Open Competition Program of Top Ten Critical Priorities of Agricultural Science and Technology Innovation for the 14th Five-Year Plan of Guangdong Province
- CARS-35/China Agriculture Research System of Ministry of Finance and Ministry of Agriculture and Rural Affairs
- CAAS-CSLPDCP-202302/Project of National Center of Technology Innovation for Pigs (NCTIP-XD/C03), and Innovation Program of Chinese Academy of Agricultural Sciences
- CAAS-ASTIP-2024-LVRI/Project of National Center of Technology Innovation for Pigs (NCTIP-XD/C03), and Innovation Program of Chinese Academy of Agricultural Sciences
LinkOut - more resources
Full Text Sources

