DNMT3B splicing dysregulation mediated by SMCHD1 loss contributes to DUX4 overexpression and FSHD pathogenesis
- PMID: 38809976
- PMCID: PMC11135424
- DOI: 10.1126/sciadv.adn7732
DNMT3B splicing dysregulation mediated by SMCHD1 loss contributes to DUX4 overexpression and FSHD pathogenesis
Abstract
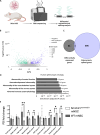
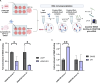
Structural maintenance of chromosomes flexible hinge domain-containing 1 (SMCHD1) is a noncanonical SMC protein and an epigenetic regulator. Mutations in SMCHD1 cause facioscapulohumeral muscular dystrophy (FSHD), by overexpressing DUX4 in muscle cells. Here, we demonstrate that SMCHD1 is a key regulator of alternative splicing in various cell types. We show how SMCHD1 loss causes splicing alterations of DNMT3B, which can lead to hypomethylation and DUX4 overexpression. Analyzing RNA sequencing data from muscle biopsies of patients with FSHD and Smchd1 knocked out cells, we found mis-splicing of hundreds of genes upon SMCHD1 loss. We conducted a high-throughput screen of splicing factors, revealing the involvement of the splicing factor RBM5 in the mis-splicing of DNMT3B. Subsequent RNA immunoprecipitation experiments confirmed that SMCHD1 is required for RBM5 recruitment. Last, we show that mis-splicing of DNMT3B leads to hypomethylation of the D4Z4 region and to DUX4 overexpression. These results suggest that DNMT3B mis-splicing due to SMCHD1 loss plays a major role in FSHD pathogenesis.
Figures







References
-
- Yoshida K., Ogawa S., Splicing factor mutations and cancer. Wiley Interdiscip. Rev. RNA 5, 445–459 (2014). - PubMed
-
- Naftelberg S., Schor I. E., Ast G., Kornblihtt A. R., Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 84, 165–198 (2015). - PubMed
-
- Dujardin G., Lafaille C., de la Mata M., Marasco L. E., Muñoz M. J., Jossic-Corcos C. L., Corcos L., Kornblihtt A. R., How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell 54, 683–690 (2014). - PubMed
-
- Ruiz-Velasco M., Kumar M., Lai M. C., Bhat P., Solis-Pinson A. B., Reyes A., Kleinsorg S., Noh K.-M., Gibson T. J., Zaugg J. B., CTCF-mediated chromatin loops between promoter and gene body regulate alternative splicing across individuals. Cell Syst. 5, 628–637.e6 (2017). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

