Rejuvenation of peripheral immune cells attenuates Alzheimer's disease-like pathologies and behavioral deficits in a mouse model
- PMID: 38809977
- PMCID: PMC11135428
- DOI: 10.1126/sciadv.adl1123
Rejuvenation of peripheral immune cells attenuates Alzheimer's disease-like pathologies and behavioral deficits in a mouse model
Abstract
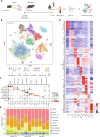
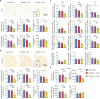
Immunosenescence contributes to systematic aging and plays a role in the pathogenesis of Alzheimer's disease (AD). Therefore, the objective of this study was to investigate the potential of immune rejuvenation as a therapeutic strategy for AD. To achieve this, the immune systems of aged APP/PS1 mice were rejuvenated through young bone marrow transplantation (BMT). Single-cell RNA sequencing revealed that young BMT restored the expression of aging- and AD-related genes in multiple cell types within blood immune cells. The level of circulating senescence-associated secretory phenotype proteins was decreased following young BMT. Notably, young BMT resulted in a significant reduction in cerebral Aβ plaque burden, neuronal degeneration, neuroinflammation, and improvement of behavioral deficits in aged APP/PS1 mice. The ameliorated cerebral amyloidosis was associated with an enhanced Aβ clearance of peripheral monocytes. In conclusion, our study provides evidence that immune system rejuvenation represents a promising therapeutic approach for AD.
Figures




Comment in
-
Immune rejuvenation - a potential AD therapy.Nat Rev Neurol. 2024 Aug;20(8):456. doi: 10.1038/s41582-024-00996-2. Nat Rev Neurol. 2024. PMID: 38977882 No abstract available.
References
-
- Sims R., Hill M., Williams J., The multiplex model of the genetics of Alzheimer's disease. Nat. Neurosci. 23, 311–322 (2020). - PubMed
-
- Bellenguez C., Küçükali F., Jansen I. E., Kleineidam L., Moreno-Grau S., Amin N., Naj A. C., Campos-Martin R., Grenier-Boley B., Andrade V., Holmans P. A., Boland A., Damotte V., van der Lee S. J., Costa M. R., Kuulasmaa T., Yang Q., de Rojas I., Bis J. C., Yaqub A., Prokic I., Chapuis J., Ahmad S., Giedraitis V., Aarsland D., Garcia-Gonzalez P., Abdelnour C., Alarcón-Martín E., Alcolea D., Alegret M., Alvarez I., Álvarez V., Armstrong N. J., Tsolaki A., Antúnez C., Appollonio I., Arcaro M., Archetti S., Pastor A. A., Arosio B., Athanasiu L., Bailly H., Banaj N., Baquero M., Barral S., Beiser A., Pastor A. B., Below J. E., Benchek P., Benussi L., Berr C., Besse C., Bessi V., Binetti G., Bizarro A., Blesa R., Boada M., Boerwinkle E., Borroni B., Boschi S., Bossù P., Bråthen G., Bressler J., Bresner C., Brodaty H., Brookes K. J., Brusco L. I., Buiza-Rueda D., Bûrger K., Burholt V., Bush W. S., Calero M., Cantwell L. B., Chene G., Chung J., Cuccaro M. L., Carracedo Á., Cecchetti R., Cervera-Carles L., Charbonnier C., Chen H. H., Chillotti C., Ciccone S., Claassen J., Clark C., Conti E., Corma-Gómez A., Costantini E., Custodero C., Daian D., Dalmasso M. C., Daniele A., Dardiotis E., Dartigues J. F., de Deyn P. P., de Paiva Lopes K., de Witte L. D., Debette S., Deckert J., Del Ser T., Denning N., DeStefano A., Dichgans M., Diehl-Schmid J., Diez-Fairen M., Rossi P. D., Djurovic S., Duron E., Düzel E., Dufouil C., Eiriksdottir G., Engelborghs S., Escott-Price V., Espinosa A., Ewers M., Faber K. M., Fabrizio T., Nielsen S. F., Fardo D. W., Farotti L., Fenoglio C., Fernández-Fuertes M., Ferrari R., Ferreira C. B., Ferri E., Fin B., Fischer P., Fladby T., Fließbach K., Fongang B., Fornage M., Fortea J., Foroud T. M., Fostinelli S., Fox N. C., Franco-Macías E., Bullido M. J., Frank-García A., Froelich L., Fulton-Howard B., Galimberti D., García-Alberca J. M., García-González P., Garcia-Madrona S., Garcia-Ribas G., Ghidoni R., Giegling I., Giorgio G., Goate A. M., Goldhardt O., Gomez-Fonseca D., González-Pérez A., Graff C., Grande G., Green E., Grimmer T., Grünblatt E., Grunin M., Gudnason V., Guetta-Baranes T., Haapasalo A., Hadjigeorgiou G., Haines J. L., Hamilton-Nelson K. L., Hampel H., Hanon O., Hardy J., Hartmann A. M., Hausner L., Harwood J., Heilmann-Heimbach S., Helisalmi S., Heneka M. T., Hernández I., Herrmann M. J., Hoffmann P., Holmes C., Holstege H., Vilas R. H., Hulsman M., Humphrey J., Biessels G. J., Jian X., Johansson C., Jun G. R., Kastumata Y., Kauwe J., Kehoe P. G., Kilander L., Ståhlbom A. K., Kivipelto M., Koivisto A., Kornhuber J., Kosmidis M. H., Kukull W. A., Kuksa P. P., Kunkle B. W., Kuzma A. B., Lage C., Laukka E. J., Launer L., Lauria A., Lee C. Y., Lehtisalo J., Lerch O., Lleó A., Longstreth W. Jr., Lopez O., de Munain A. L., Love S., Löwemark M., Luckcuck L., Lunetta K. L., Ma Y., Macías J., MacLeod C. A., Maier W., Mangialasche F., Spallazzi M., Marquié M., Marshall R., Martin E. R., Montes A. M., Rodríguez C. M., Masullo C., Mayeux R., Mead S., Mecocci P., Medina M., Meggy A., Mehrabian S., Mendoza S., Menéndez-González M., Mir P., Moebus S., Mol M., Molina-Porcel L., Montrreal L., Morelli L., Moreno F., Morgan K., Mosley T., Nöthen M. M., Muchnik C., Mukherjee S., Nacmias B., Ngandu T., Nicolas G., Nordestgaard B. G., Olaso R., Orellana A., Orsini M., Ortega G., Padovani A., Paolo C., Papenberg G., Parnetti L., Pasquier F., Pastor P., Peloso G., Pérez-Cordón A., Pérez-Tur J., Pericard P., Peters O., Pijnenburg Y. A. L., Pineda J. A., Piñol-Ripoll G., Pisanu C., Polak T., Popp J., Posthuma D., Priller J., Puerta R., Quenez O., Quintela I., Thomassen J. Q., Rábano A., Rainero I., Rajabli F., Ramakers I., Real L. M., Reinders M. J. T., Reitz C., Reyes-Dumeyer D., Ridge P., Riedel-Heller S., Riederer P., Roberto N., Rodriguez-Rodriguez E., Rongve A., Allende I. R., Rosende-Roca M., Royo J. L., Rubino E., Rujescu D., Sáez M. E., Sakka P., Saltvedt I., Sanabria Á., Sánchez-Arjona M. B., Sanchez-Garcia F., Juan P. S., Sánchez-Valle R., Sando S. B., Sarnowski C., Satizabal C. L., Scamosci M., Scarmeas N., Scarpini E., Scheltens P., Scherbaum N., Scherer M., Schmid M., Schneider A., Schott J. M., Selbæk G., Seripa D., Serrano M., Sha J., Shadrin A. A., Skrobot O., Slifer S., Snijders G. J. L., Soininen H., Solfrizzi V., Solomon A., Song Y., Sorbi S., Sotolongo-Grau O., Spalletta G., Spottke A., Squassina A., Stordal E., Tartan J. P., Tárraga L., Tesí N., Thalamuthu A., Thomas T., Tosto G., Traykov L., Tremolizzo L., Tybjærg-Hansen A., Uitterlinden A., Ullgren A., Ulstein I., Valero S., Valladares O., Broeckhoven C. V., Vance J., Vardarajan B. N., van der Lugt A., Dongen J. V., van Rooij J., van Swieten J., Vandenberghe R., Verhey F., Vidal J. S., Vogelgsang J., Vyhnalek M., Wagner M., Wallon D., Wang L. S., Wang R., Weinhold L., Wiltfang J., Windle G., Woods B., Yannakoulia M., Zare H., Zhao Y., Zhang X., Zhu C., Zulaica M., Farrer L. A., Psaty B. M., Ghanbari M., Raj T., Sachdev P., Mather K., Jessen F., Ikram M. A., de Mendonça A., Hort J., Tsolaki M., Pericak-Vance M. A., Amouyel P., Williams J., Frikke-Schmidt R., Clarimon J., Deleuze J. F., Rossi G., Seshadri S., Andreassen O. A., Ingelsson M., Hiltunen M., Sleegers K., Schellenberg G. D., van Duijn C. M., Sims R., van der Flier W. M., Ruiz A., Ramirez A., Lambert J. C., New insights into the genetic etiology of Alzheimer's disease and related dementias. Nat. Genet. 54, 412–436 (2022). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

