CMTR-1 RNA methyltransferase mutations activate widespread expression of a dopaminergic neuron-specific mitochondrial complex I gene
- PMID: 38810637
- PMCID: PMC11265314
- DOI: 10.1016/j.cub.2024.04.079
CMTR-1 RNA methyltransferase mutations activate widespread expression of a dopaminergic neuron-specific mitochondrial complex I gene
Abstract
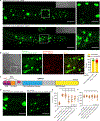
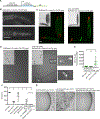
The mitochondrial proteome is comprised of approximately 1,100 proteins,1 all but 12 of which are encoded by the nuclear genome in C. elegans. The expression of nuclear-encoded mitochondrial proteins varies widely across cell lineages and metabolic states,2,3,4 but the factors that specify these programs are not known. Here, we identify mutations in two nuclear-localized mRNA processing proteins, CMTR1/CMTR-1 and SRRT/ARS2/SRRT-1, which we show act via the same mechanism to rescue the mitochondrial complex I mutant NDUFS2/gas-1(fc21). CMTR-1 is an FtsJ-family RNA methyltransferase that, in mammals, 2'-O-methylates the first nucleotide 3' to the mRNA CAP to promote RNA stability and translation5,6,7,8. The mutations isolated in cmtr-1 are dominant and lie exclusively in the regulatory G-patch domain. SRRT-1 is an RNA binding partner of the nuclear cap-binding complex and determines mRNA transcript fate.9 We show that cmtr-1 and srrt-1 mutations activate embryonic expression of NDUFS2/nduf-2.2, a paralog of NDUFS2/gas-1 normally expressed only in dopaminergic neurons, and that nduf-2.2 is necessary for the complex I rescue by the cmtr-1 G-patch mutant. Additionally, we find that loss of the cmtr-1 G-patch domain cause ectopic localization of CMTR-1 protein to processing bodies (P bodies), phase-separated organelles involved in mRNA storage and decay.10 P-body localization of the G-patch mutant CMTR-1 contributes to the rescue of the hyperoxia sensitivity of the NDUFS2/gas-1 mutant. This study suggests that mRNA methylation at P bodies may control nduf-2.2 gene expression, with broader implications for how the mitochondrial proteome is translationally remodeled in the face of tissue-specific metabolic requirements and stress.
Keywords: C. elegans; CMTR1; G-patch domain; Mitochondria; NADH:ubiquinone oxidoreductase; SRRT; complex I; hyperoxia; mRNA methylation; processing bodies.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests V.K.M. is listed as an inventor on patents filed by MGH on therapeutic uses of hypoxia. V.K.M. is a paid advisor to 5a.m. Ventures.
Figures


References
-
- Grimsrud PA, Carson JJ, Hebert AS, Hubler SL, Niemi NM, Bailey DJ, Jochem A, Stapleton DS, Keller MP, Westphall MS, et al. (2012). A Quantitative Map of the Liver Mitochondrial Phosphoproteome Reveals Posttranslational Control of Ketogenesis. Cell Metab 16, 672–683. 10.1016/j.cmet.2012.10.004. - DOI - PMC - PubMed
-
- Kuhn KM, DeRisi JL, Brown PO, and Sarnow P (2001). Global and Specific Translational Regulation in the Genomic Response of Saccharomyces cerevisiae to a Rapid Transfer from a Fermentable to a Nonfermentable Carbon Source. Mol Cell Biol 21, 916–927. 10.1128/mcb.21.3.916-927.2001. - DOI - PMC - PubMed
-
- Picard-Jean F, Brand C, Tremblay-Létourneau M, Allaire A, Beaudoin MC, Boudreault S, Duval C, Rainville-Sirois J, Robert F, Pelletier J, et al. (2018). 2’-O-methylation of the mRNA cap protects RNAs from decapping and degradation by DXO. PLoS ONE 13, e0193804. 10.1371/journal.pone.0193804. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

