Expression and characterization of a novel microbial GH9 glucanase, IDSGLUC9-4, isolated from sheep rumen
- PMID: 38810985
- PMCID: PMC11366526
- DOI: 10.5713/ab.24.0138
Expression and characterization of a novel microbial GH9 glucanase, IDSGLUC9-4, isolated from sheep rumen
Abstract
Objective: This study aimed to identify and characterize a novel endo-β-glucanase, IDSGLUC9-4, from the rumen metatranscriptome of Hu sheep.
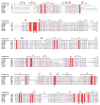
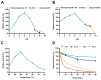
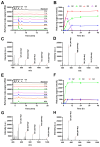
Methods: A novel endo-β-glucanase, IDSGLUC9-4, was heterologously expressed in Escherichia coli and biochemically characterized. The optimal temperature and pH of recombinant IDSGLUC9-4 were determined. Subsequently, substrate specificity of the enzyme was assessed using mixed-linked glucans including barley β-glucan and Icelandic moss lichenan. Thin-layer chromatography (TLC), high-performance liquid chromatography (HPLC), matrix assisted laser desorption ionization time of flight mass spectrometry analyses were conducted to determine the products released from polysaccharides and cello-oligosaccharides substrates.
Results: The recombinant IDSGLUC9-4 exhibited temperature and pH optima of 40°C and pH 6.0, respectively. It exclusively hydrolyzed mixed-linked glucans, with significant activity observed for barley β-glucan (109.59±3.61 μmol/mg min) and Icelandic moss lichenan (35.35±1.55 μmol/mg min). TLC and HPLC analyses revealed that IDSGLUC9-4 primarily released cellobiose, cellotriose, and cellotetraose from polysaccharide substrates. Furthermore, after 48 h of reaction, IDSGLUC9-4 removed most of the glucose, indicating transglycosylation activity alongside its endo-glucanase activity.
Conclusion: The recombinant IDSGLUC9-4 was a relatively acid-resistant, mesophilic endo-glucanase (EC 3.2.1.4) that hydrolyzed glucan-like substrates, generating predominantly G3 and G4 oligosaccharides, and which appeared to have glycosylation activity. These findings provided insights into the substrate specificity and product profiles of rumen-derived GH9 glucanases and contributed to the expanding knowledge of cellulolytic enzymes and novel herbivore rumen enzymes in general.
Keywords: Expression; Glycoside Hydrolase; Hydrolysis; Rumen Microbiome; β-Glucan.
Conflict of interest statement
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Figures




References
-
- Beckmann L, Simon O, Vahjen W. Isolation and identification of mixed linked beta -glucan degrading bacteria in the intestine of broiler chickens and partial characterization of respective 1,3-1,4-beta-glucanase activities. J Basic Microbiol. 2006;46:175–85. doi: 10.1002/jobm.200510107. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

