Stochasticity, determinism, and contingency shape genome evolution of endosymbiotic bacteria
- PMID: 38811551
- PMCID: PMC11137140
- DOI: 10.1038/s41467-024-48784-2
Stochasticity, determinism, and contingency shape genome evolution of endosymbiotic bacteria
Abstract
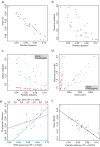
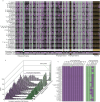
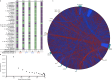
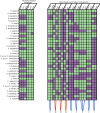
Evolution results from the interaction of stochastic and deterministic processes that create a web of historical contingency, shaping gene content and organismal function. To understand the scope of this interaction, we examine the relative contributions of stochasticity, determinism, and contingency in shaping gene inactivation in 34 lineages of endosymbiotic bacteria, Sodalis, found in parasitic lice, Columbicola, that are independently undergoing genome degeneration. Here we show that the process of genome degeneration in this system is largely deterministic: genes involved in amino acid biosynthesis are lost while those involved in providing B-vitamins to the host are retained. In contrast, many genes encoding redundant functions, including components of the respiratory chain and DNA repair pathways, are subject to stochastic loss, yielding historical contingencies that constrain subsequent losses. Thus, while selection results in functional convergence between symbiont lineages, stochastic mutations initiate distinct evolutionary trajectories, generating diverse gene inventories that lack the functional redundancy typically found in free-living relatives.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Ongoing Transposon-Mediated Genome Reduction in the Luminous Bacterial Symbionts of Deep-Sea Ceratioid Anglerfishes.mBio. 2018 Jun 26;9(3):e01033-18. doi: 10.1128/mBio.01033-18. mBio. 2018. PMID: 29946051 Free PMC article.
-
Differential genome evolution between companion symbionts in an insect-bacterial symbiosis.mBio. 2014 Sep 30;5(5):e01697-14. doi: 10.1128/mBio.01697-14. mBio. 2014. PMID: 25271287 Free PMC article.
-
Chance and necessity in the genome evolution of endosymbiotic bacteria of insects.ISME J. 2017 Jun;11(6):1291-1304. doi: 10.1038/ismej.2017.18. Epub 2017 Mar 21. ISME J. 2017. PMID: 28323281 Free PMC article.
-
In it for the long haul: evolutionary consequences of persistent endosymbiosis.Curr Opin Genet Dev. 2017 Dec;47:83-90. doi: 10.1016/j.gde.2017.08.006. Epub 2017 Sep 19. Curr Opin Genet Dev. 2017. PMID: 28934627 Review.
-
Taxonomy of lice and their endosymbiotic bacteria in the post-genomic era.Clin Microbiol Infect. 2012 Apr;18(4):324-31. doi: 10.1111/j.1469-0691.2012.03782.x. Clin Microbiol Infect. 2012. PMID: 22429457 Review.
Cited by
-
A systematic framework for understanding the microbiome in human health and disease: from basic principles to clinical translation.Signal Transduct Target Ther. 2024 Sep 23;9(1):237. doi: 10.1038/s41392-024-01946-6. Signal Transduct Target Ther. 2024. PMID: 39307902 Free PMC article. Review.
-
Effect of Oceanic Islands on an Insect Symbiont Genome in Transition to a Host-Restricted Lifestyle.Genome Biol Evol. 2025 Jul 30;17(8):evaf153. doi: 10.1093/gbe/evaf153. Genome Biol Evol. 2025. PMID: 40711997 Free PMC article.
References
-
- Buchner, P. Endosymbiosis of animals with plant microorganisms. 1–909 (Interscience, 1965).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

