Small molecule induced STING degradation facilitated by the HECT ligase HERC4
- PMID: 38811577
- PMCID: PMC11137104
- DOI: 10.1038/s41467-024-48922-w
Small molecule induced STING degradation facilitated by the HECT ligase HERC4
Abstract
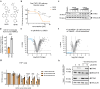
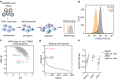
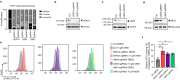
Stimulator of interferon genes (STING) is a central component of the cytosolic nucleic acids sensing pathway and as such master regulator of the type I interferon response. Due to its critical role in physiology and its' involvement in a variety of diseases, STING has been a focus for drug discovery. Targeted protein degradation (TPD) has emerged as a promising pharmacology for targeting previously considered undruggable proteins by hijacking the cellular ubiquitin proteasome system (UPS) with small molecules. Here, we identify AK59 as a STING degrader leveraging HERC4, a HECT-domain E3 ligase. Additionally, our data reveals that AK59 is effective on the common pathological STING mutations, suggesting a potential clinical application of this mechanism. Thus, these findings introduce HERC4 to the fields of TPD and of compound-induced degradation of STING, suggesting potential therapeutic applications.
© 2024. The Author(s).
Conflict of interest statement
M.M., I.S., B.G., F.F., D.B., O.S., N.P., E.A., S.K., S.R., D.J., T.T., R.M., S.A., A.R., F.N., A.H., C.N.P., and D.G. are current employees and shareholders of Novartis Pharma. A.I.M., D.Bo., and J.R.H. are former employees of Novartis Pharma. D.Bo. is a current employee of Monte Rosa Therapeutics, Basel, Switzerland., J.R.H. is a current employee of Vector Biology, Cambridge, MA, USA.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

