A Gram-negative-selective antibiotic that spares the gut microbiome
- PMID: 38811738
- PMCID: PMC12135874
- DOI: 10.1038/s41586-024-07502-0
A Gram-negative-selective antibiotic that spares the gut microbiome
Abstract
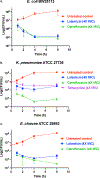
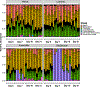
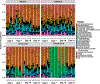
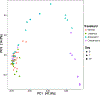
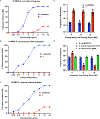
Infections caused by Gram-negative pathogens are increasingly prevalent and are typically treated with broad-spectrum antibiotics, resulting in disruption of the gut microbiome and susceptibility to secondary infections1-3. There is a critical need for antibiotics that are selective both for Gram-negative bacteria over Gram-positive bacteria, as well as for pathogenic bacteria over commensal bacteria. Here we report the design and discovery of lolamicin, a Gram-negative-specific antibiotic targeting the lipoprotein transport system. Lolamicin has activity against a panel of more than 130 multidrug-resistant clinical isolates, shows efficacy in multiple mouse models of acute pneumonia and septicaemia infection, and spares the gut microbiome in mice, preventing secondary infection with Clostridioides difficile. The selective killing of pathogenic Gram-negative bacteria by lolamicin is a consequence of low sequence homology for the target in pathogenic bacteria versus commensals; this doubly selective strategy can be a blueprint for the development of other microbiome-sparing antibiotics.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures



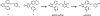
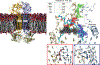

Comment in
-
A selective microbiome-sparing antibiotic.Nat Rev Drug Discov. 2024 Aug;23(8):581. doi: 10.1038/d41573-024-00113-4. Nat Rev Drug Discov. 2024. PMID: 38937617 No abstract available.
References
Main Text References
-
- Lynch SV & Pedersen O The Human Intestinal Microbiome in Health and Disease. New Eng J Med 375, 2369–2379 (2016). - PubMed
-
- Owens RC Jr., Donskey CJ, Gaynes RP, Loo VG & Muto CA Antimicrobial-associated risk factors for Clostridium difficile infection. Clin Infect Dis 46, S19–31 (2008). - PubMed
-
- Iizumi T, Battaglia T, Ruiz V & Perez Perez GI Gut Microbiome and Antibiotics. Arch Med Res 48, 727–734 (2017). - PubMed
Additional References:
-
- Pandit KR & Klauda JB Membrane models of E. coli containing cyclic moieties in the aliphatic lipid chain. Biochim Biophys Acta 1818, 1205–1210 (2012). - PubMed
-
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW & Klein ML Comparison of simple potential functions for simulating liquid water. J Chem Phys 79, 926–935 (1983).
-
- Jo S, Kim T, Iyer VG & Im W CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29, 1859–1865 (2008). - PubMed
-
- Humphrey W, Dalke A & Schulten K VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38 (1996). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

