The sex of organ geometry
- PMID: 38811741
- PMCID: PMC11168936
- DOI: 10.1038/s41586-024-07463-4
The sex of organ geometry
Abstract
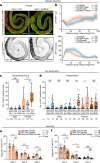
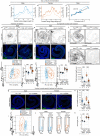
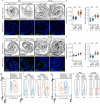
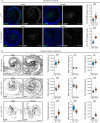
Organs have a distinctive yet often overlooked spatial arrangement in the body1-5. We propose that there is a logic to the shape of an organ and its proximity to its neighbours. Here, by using volumetric scans of many Drosophila melanogaster flies, we develop methods to quantify three-dimensional features of organ shape, position and interindividual variability. We find that both the shapes of organs and their relative arrangement are consistent yet differ between the sexes, and identify unexpected interorgan adjacencies and left-right organ asymmetries. Focusing on the intestine, which traverses the entire body, we investigate how sex differences in three-dimensional organ geometry arise. The configuration of the adult intestine is only partially determined by physical constraints imposed by adjacent organs; its sex-specific shape is actively maintained by mechanochemical crosstalk between gut muscles and vascular-like trachea. Indeed, sex-biased expression of a muscle-derived fibroblast growth factor-like ligand renders trachea sexually dimorphic. In turn, tracheal branches hold gut loops together into a male or female shape, with physiological consequences. Interorgan geometry represents a previously unrecognized level of biological complexity which might enable or confine communication across organs and could help explain sex or species differences in organ function.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- Aristotle. The History of Animals (Zinc Read, 2023).
-
- Falcon, A. & Lefebvre, D. Aristotle’s Generation of Animals (Cambridge Univ. Press, 2018).
-
- Hippocrates of Kos & Adams, F. On the Sacred Disease (Dalcassian, 2023).
-
- Leroi, A. M. The Lagoon: How Aristotle Invented Science (Bloomsbury, 2015).
-
- Vesalius, A. The Fabric of the Human Body: An Annotated Translation of the 1543 and 1555 Editions of “De Humani Corporis Fabrica Libri Septem”, by D.H. Garrison and M.H. Hast (Karger, 2013).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

