MYC phase separation selectively modulates the transcriptome
- PMID: 38811792
- PMCID: PMC11479839
- DOI: 10.1038/s41594-024-01322-6
MYC phase separation selectively modulates the transcriptome
Erratum in
-
Author Correction: MYC phase separation selectively modulates the transcriptome.Nat Struct Mol Biol. 2024 Nov;31(11):1808. doi: 10.1038/s41594-024-01351-1. Nat Struct Mol Biol. 2024. PMID: 38867115 No abstract available.
Abstract
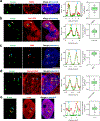
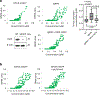
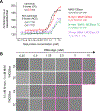
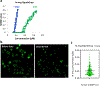
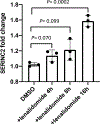
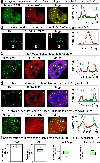
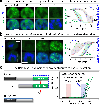
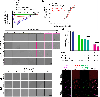
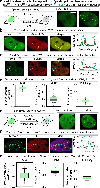
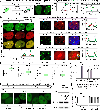
Dysregulation and enhanced expression of MYC transcription factors (TFs) including MYC and MYCN contribute to the majority of human cancers. For example, MYCN is amplified up to several hundredfold in high-risk neuroblastoma. The resulting overexpression of N-myc aberrantly activates genes that are not activated at low N-myc levels and drives cell proliferation. Whether increasing N-myc levels simply mediates binding to lower-affinity binding sites in the genome or fundamentally changes the activation process remains unclear. One such activation mechanism that could become important above threshold levels of N-myc is the formation of aberrant transcriptional condensates through phase separation. Phase separation has recently been linked to transcriptional regulation, but the extent to which it contributes to gene activation remains an open question. Here we characterized the phase behavior of N-myc and showed that it can form dynamic condensates that have transcriptional hallmarks. We tested the role of phase separation in N-myc-regulated transcription by using a chemogenetic tool that allowed us to compare non-phase-separated and phase-separated conditions at equivalent N-myc levels, both of which showed a strong impact on gene expression compared to no N-myc expression. Interestingly, we discovered that only a small percentage (<3%) of N-myc-regulated genes is further modulated by phase separation but that these events include the activation of key oncogenes and the repression of tumor suppressors. Indeed, phase separation increases cell proliferation, corroborating the biological effects of the transcriptional changes. However, our results also show that >97% of N-myc-regulated genes are not affected by N-myc phase separation, demonstrating that soluble complexes of TFs with the transcriptional machinery are sufficient to activate transcription.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Figures

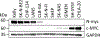




References
Methods-only Reference
-
- Pillai-Kastoori L, Schutz-Geschwender AR & Harford JA A systematic approach to quantitative Western blot analysis. Analytical Biochemistry 593, 113608 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

