H3.3-G34W in giant cell tumor of bone functionally aligns with the exon choice repressor hnRNPA1L2
- PMID: 38811797
- PMCID: PMC11327103
- DOI: 10.1038/s41417-024-00776-6
H3.3-G34W in giant cell tumor of bone functionally aligns with the exon choice repressor hnRNPA1L2
Erratum in
-
Correction: H3.3-G34W in giant cell tumor of bone functionally aligns with the exon choice repressor hnRNPA1L2.Cancer Gene Ther. 2024 Aug;31(8):1281. doi: 10.1038/s41417-024-00803-6. Cancer Gene Ther. 2024. PMID: 38937572 Free PMC article. No abstract available.
Abstract
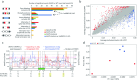
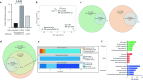
RNA processing is an essential post-transcriptional phenomenon that provides the necessary complexity of transcript diversity prior to translation. Aberrations in this process could contribute to tumourigenesis, and we have previously reported increased splicing alterations in giant cell tumor of bone (GCTB), which carries mutations in the histone variant H3.3 encoding glycine 34 substituted for tryptophan (H3.3-G34W). G34W interacts with several splicing factors, most notably the trans-acting splicing factor hnRNPA1L2. To gain a deeper understanding of RNA processing in GCTB and isogenic HeLa cells with H3.3-G34W, we generated RNA-immunoprecipitation sequencing data from hnRNPA1L2 and H3.3-G34W associated RNAs, which showed that 80% overlapped across genic regions and were frequently annotated as E2F transcription factor binding sites. Splicing aberrations in both GCTB and HeLa cells with H3.3-G34W were significantly enriched for known hnRNPA1L2 binding motifs (p value < 0.01). This splicing aberration differed from hnRNPA1L2 knockouts, which showed alterations independent of H3.3-G34W. Of functional significance, hnRNPA1L2 was redistributed to closely match the H3.3 pattern, likely driven by G34W, and to loci not occupied in normal parental cells. Taken together, our data reveal a functional overlap between hnRNPA1L2 and H3.3-G34W with likely significant consequences for RNA processing during GCTB pathogenesis. This provides novel opportunities for therapeutic intervention in future modus operandi.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
The histone variant H3.3 G34W substitution in giant cell tumor of the bone link chromatin and RNA processing.Sci Rep. 2017 Oct 18;7(1):13459. doi: 10.1038/s41598-017-13887-y. Sci Rep. 2017. PMID: 29044188 Free PMC article.
-
Significance of Histone H3.3 (G34W)-Mutant Protein in Pathological Diagnosis of Giant Cell Tumor of Bone.Asian Pac J Cancer Prev. 2023 May 1;24(5):1737-1741. doi: 10.31557/APJCP.2023.24.5.1737. Asian Pac J Cancer Prev. 2023. PMID: 37247296 Free PMC article.
-
H3F3A (Histone 3.3) G34W Immunohistochemistry: A Reliable Marker Defining Benign and Malignant Giant Cell Tumor of Bone.Am J Surg Pathol. 2017 Aug;41(8):1059-1068. doi: 10.1097/PAS.0000000000000859. Am J Surg Pathol. 2017. PMID: 28505000 Free PMC article.
-
Histone H3.3 mutation in giant cell tumor of bone: an update in pathology.Med Mol Morphol. 2020 Mar;53(1):1-6. doi: 10.1007/s00795-019-00238-1. Epub 2019 Nov 20. Med Mol Morphol. 2020. PMID: 31748824 Review.
-
Giant cell tumor of bone: updated molecular pathogenesis and tumor biology.Hum Pathol. 2018 Nov;81:1-8. doi: 10.1016/j.humpath.2018.06.017. Epub 2018 Jun 23. Hum Pathol. 2018. PMID: 29944971 Review.
Cited by
-
Emerging roles of cancer-associated histone mutations in genomic instabilities.Front Cell Dev Biol. 2024 Oct 8;12:1455572. doi: 10.3389/fcell.2024.1455572. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39439908 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

