Variant-specific pathophysiological mechanisms of AFF3 differently influence transcriptome profiles
- PMID: 38811945
- PMCID: PMC11137988
- DOI: 10.1186/s13073-024-01339-y
Variant-specific pathophysiological mechanisms of AFF3 differently influence transcriptome profiles
Abstract
Background: We previously described the KINSSHIP syndrome, an autosomal dominant disorder associated with intellectual disability (ID), mesomelic dysplasia and horseshoe kidney, caused by de novo variants in the degron of AFF3. Mouse knock-ins and overexpression in zebrafish provided evidence for a dominant-negative mode of action, wherein an increased level of AFF3 resulted in pathological effects.
Methods: Evolutionary constraints suggest that other modes-of-inheritance could be at play. We challenged this hypothesis by screening ID cohorts for individuals with predicted-to-be damaging variants in AFF3. We used both animal and cellular models to assess the deleteriousness of the identified variants.
Results: We identified an individual with a KINSSHIP-like phenotype carrying a de novo partial duplication of AFF3 further strengthening the hypothesis that an increased level of AFF3 is pathological. We also detected seventeen individuals displaying a milder syndrome with either heterozygous Loss-of-Function (LoF) or biallelic missense variants in AFF3. Consistent with semi-dominance, we discovered three patients with homozygous LoF and one compound heterozygote for a LoF and a missense variant, who presented more severe phenotypes than their heterozygous parents. Matching zebrafish knockdowns exhibit neurological defects that could be rescued by expressing human AFF3 mRNA, confirming their association with the ablation of aff3. Conversely, some of the human AFF3 mRNAs carrying missense variants identified in affected individuals did not rescue these phenotypes. Overexpression of mutated AFF3 mRNAs in zebrafish embryos produced a significant increase of abnormal larvae compared to wild-type overexpression further demonstrating deleteriousness. To further assess the effect of AFF3 variation, we profiled the transcriptome of fibroblasts from affected individuals and engineered isogenic cells harboring + / + , KINSSHIP/KINSSHIP, LoF/ + , LoF/LoF or KINSSHIP/LoF AFF3 genotypes. The expression of more than a third of the AFF3 bound loci is modified in either the KINSSHIP/KINSSHIP or the LoF/LoF lines. While the same pathways are affected, only about one third of the differentially expressed genes are common to the homozygote datasets, indicating that AFF3 LoF and KINSSHIP variants largely modulate transcriptomes differently, e.g. the DNA repair pathway displayed opposite modulation.
Conclusions: Our results and the high pleiotropy shown by variation at this locus suggest that minute changes in AFF3 function are deleterious.
Keywords: Horseshoe kidney; Intellectual disability; Mesomelic dysplasia; Transcriptome; Zebrafish model.
© 2024. The Author(s).
Conflict of interest statement
Annabelle Tuttle, Houda Zghal Elloumi and Chaofan Zhang are employees of GeneDx and Desiree DeMille works for ARUP Laboratories. James R. Lupski has stock ownership in 23andMe and is a paid consultant for Genome International. Claudia M.B. Carvalho provides consulting service for Ionis Pharmaceuticals. The other authors have no competing interests to declare.
Figures
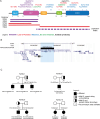

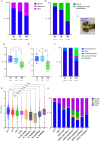

Update of
-
Variant-specific pathophysiological mechanisms of AFF3 differently influence transcriptome profiles.medRxiv [Preprint]. 2024 Jan 17:2024.01.14.24301100. doi: 10.1101/2024.01.14.24301100. medRxiv. 2024. Update in: Genome Med. 2024 May 30;16(1):72. doi: 10.1186/s13073-024-01339-y. PMID: 38293053 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- WT223718/Z/21/Z/WT_/Wellcome Trust/United Kingdom
- U01 HG011758/HG/NHGRI NIH HHS/United States
- 31003A_182632/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung
- R35 NS105078/NS/NINDS NIH HHS/United States
- IZSTZ0_216615/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

