JUN mediates the senescence associated secretory phenotype and immune cell recruitment to prevent prostate cancer progression
- PMID: 38811984
- PMCID: PMC11134959
- DOI: 10.1186/s12943-024-02022-x
JUN mediates the senescence associated secretory phenotype and immune cell recruitment to prevent prostate cancer progression
Abstract
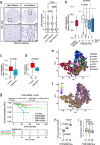
Background: Prostate cancer develops through malignant transformation of the prostate epithelium in a stepwise, mutation-driven process. Although activator protein-1 transcription factors such as JUN have been implicated as potential oncogenic drivers, the molecular programs contributing to prostate cancer progression are not fully understood.
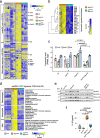
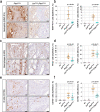
Methods: We analyzed JUN expression in clinical prostate cancer samples across different stages and investigated its functional role in a Pten-deficient mouse model. We performed histopathological examinations, transcriptomic analyses and explored the senescence-associated secretory phenotype in the tumor microenvironment.
Results: Elevated JUN levels characterized early-stage prostate cancer and predicted improved survival in human and murine samples. Immune-phenotyping of Pten-deficient prostates revealed high accumulation of tumor-infiltrating leukocytes, particularly innate immune cells, neutrophils and macrophages as well as high levels of STAT3 activation and IL-1β production. Jun depletion in a Pten-deficient background prevented immune cell attraction which was accompanied by significant reduction of active STAT3 and IL-1β and accelerated prostate tumor growth. Comparative transcriptome profiling of prostate epithelial cells revealed a senescence-associated gene signature, upregulation of pro-inflammatory processes involved in immune cell attraction and of chemokines such as IL-1β, TNF-α, CCL3 and CCL8 in Pten-deficient prostates. Strikingly, JUN depletion reversed both the senescence-associated secretory phenotype and senescence-associated immune cell infiltration but had no impact on cell cycle arrest. As a result, JUN depletion in Pten-deficient prostates interfered with the senescence-associated immune clearance and accelerated tumor growth.
Conclusions: Our results suggest that JUN acts as tumor-suppressor and decelerates the progression of prostate cancer by transcriptional regulation of senescence- and inflammation-associated genes. This study opens avenues for novel treatment strategies that could impede disease progression and improve patient outcomes.
Keywords: AP-1 transcription factors; Immune infiltration; JUN; Prostate cancer; SASP; Senescence.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

