Depression proteomic profiling in adolescents with transcriptome analyses in independent cohorts
- PMID: 38812487
- PMCID: PMC11133714
- DOI: 10.3389/fpsyt.2024.1372106
Depression proteomic profiling in adolescents with transcriptome analyses in independent cohorts
Abstract
Introduction: Depression is a major global burden with unclear pathophysiology and poor treatment outcomes. Diagnosis of depression continues to rely primarily on behavioral rather than biological methods. Investigating tools that might aid in diagnosing and treating early-onset depression is essential for improving the prognosis of the disease course. While there is increasing evidence of possible biomarkers in adult depression, studies investigating this subject in adolescents are lacking.
Methods: In the current study, we analyzed protein levels in 461 adolescents assessed for depression using the Development and Well-Being Assessment (DAWBA) questionnaire as part of the domestic Psychiatric Health in Adolescent Study conducted in Uppsala, Sweden. We used the Proseek Multiplex Neuro Exploratory panel with Proximity Extension Assay technology provided by Olink Bioscience, followed by transcriptome analyses for the genes corresponding to the significant proteins, using four publicly available cohorts.
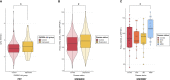
Results: We identified a total of seven proteins showing different levels between DAWBA risk groups at nominal significance, including RBKS, CRADD, ASGR1, HMOX2, PPP3R1, CD63, and PMVK. Transcriptomic analyses for these genes showed nominally significant replication of PPP3R1 in two of four cohorts including whole blood and prefrontal cortex, while ASGR1 and CD63 were replicated in only one cohort.
Discussion: Our study on adolescent depression revealed protein-level and transcriptomic differences, particularly in PPP3R1, pointing to the involvement of the calcineurin pathway in depression. Our findings regarding PPP3R1 also support the role of the prefrontal cortex in depression and reinforce the significance of investigating prefrontal cortex-related mechanisms in depression.
Keywords: adolescents; depression; proteome; psychiatry; transcriptome.
Copyright © 2024 Sokolov, Lafta, Nordberg, Jonsson and Schiöth.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Circulating proteins as predictors of cardiovascular mortality in end-stage renal disease.J Nephrol. 2019 Feb;32(1):111-119. doi: 10.1007/s40620-018-0556-5. Epub 2018 Nov 29. J Nephrol. 2019. PMID: 30499038 Free PMC article.
-
Biomarkers in Child and Adolescent Depression.Child Psychiatry Hum Dev. 2023 Feb;54(1):266-281. doi: 10.1007/s10578-021-01246-y. Epub 2021 Sep 29. Child Psychiatry Hum Dev. 2023. PMID: 34590201 Free PMC article. Review.
-
Identification of Plasma Inflammatory Markers of Adolescent Depression Using the Olink Proteomics Platform.J Inflamm Res. 2023 Oct 11;16:4489-4501. doi: 10.2147/JIR.S425780. eCollection 2023. J Inflamm Res. 2023. PMID: 37849645 Free PMC article.
-
High levels of cerebrospinal fluid chemokines point to the presence of neuroinflammation in peripheral neuropathic pain: a cross-sectional study of 2 cohorts of patients compared with healthy controls.Pain. 2017 Dec;158(12):2487-2495. doi: 10.1097/j.pain.0000000000001061. Pain. 2017. PMID: 28930774 Free PMC article.
-
Retrospective analysis: reproducibility of interblastomere differences of mRNA expression in 2-cell stage mouse embryos is remarkably poor due to combinatorial mechanisms of blastomere diversification.Mol Hum Reprod. 2018 Jul 1;24(7):388-400. doi: 10.1093/molehr/gay021. Mol Hum Reprod. 2018. PMID: 29746690
Cited by
-
Multi-omics Analysis of Energy Metabolism Pathways Across Major Psychiatric Disorders.Mol Neurobiol. 2025 Jun 11. doi: 10.1007/s12035-025-05133-8. Online ahead of print. Mol Neurobiol. 2025. PMID: 40498271
References
LinkOut - more resources
Full Text Sources
Miscellaneous

