Rapid spread of the SARS-CoV-2 JN.1 lineage is associated with increased neutralization evasion
- PMID: 38812550
- PMCID: PMC11134884
- DOI: 10.1016/j.isci.2024.109904
Rapid spread of the SARS-CoV-2 JN.1 lineage is associated with increased neutralization evasion
Abstract
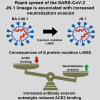
In July/August 2023, the highly mutated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) BA.2.86 lineage emerged and its descendant JN.1 is on track to become the dominant SARS-CoV-2 lineage globally. Compared to the spike (S) protein of the parental BA.2.86 lineage, the JN.1 S protein contains one mutation, L455S, which may affect receptor binding and antibody evasion. Here, we performed a virological assessment of the JN.1 lineage employing pseudovirus particles bearing diverse SARS-CoV-2 S proteins. Using this strategy, it was found that S protein mutation L455S confers increased neutralization resistance but reduces ACE2 binding capacity and S protein-driven cell entry efficiency. Altogether, these data suggest that the benefit of increased antibody evasion outweighs the reduced ACE2 binding capacity and further enabled the JN.1 lineage to effectively spread in the human population.
Keywords: Health sciences; Immunology; Virology.
© 2024 The Author(s).
Conflict of interest statement
S.P. and M.H. conducted contract research (testing of vaccinee sera for neutralizing activity against SARS-CoV-2) for Valneva unrelated to this work. A.D.-J. served as advisor for Pfizer, unrelated to this work. G.M.N.B. served as advisor for Moderna, unrelated to this work. S.P. served as advisor for BioNTech, unrelated to this work.
Figures


References
-
- Zhang L., Kempf A., Nehlmeier I., Cossmann A., Richter A., Bdeir N., Graichen L., Moldenhauer A.S., Dopfer-Jablonka A., Stankov M.V., et al. SARS-CoV-2 BA.2.86 enters lung cells and evades neutralizing antibodies with high efficiency. Cell. 2024;187:596–608.e17. doi: 10.1016/j.cell.2023.12.025. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

