Improvements of weaned pigs barn hygiene to reduce the spread of antimicrobial resistance
- PMID: 38812683
- PMCID: PMC11135127
- DOI: 10.3389/fmicb.2024.1393923
Improvements of weaned pigs barn hygiene to reduce the spread of antimicrobial resistance
Abstract
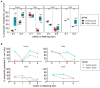
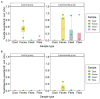
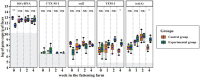
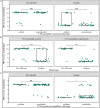
The spread of antimicrobial resistance (AMR) in animal husbandry is usually attributed to the use of antibiotics and poor hygiene and biosecurity. We therefore conducted experimental trials to improve hygiene management in weaned pig houses and assessed the impact on the spread. For each of the two groups examined, the experimental group (EG) and the control group (CG), three replicate batches of piglets from the same pig breeder, kept in pre-cleaned flat decks, were analyzed. In the flat decks of the experimental groups, the hygiene conditions (cleaning, disinfection, dust removal and fly control) were improved, while regular hygiene measures were carried out in the control groups. The occurrence and spread of AMR were determined in Escherichia coli (E. coli; resistance indicator) using cultivation-dependent (CFU) and -independent (qPCR) methods as well as whole genome sequencing of isolates in samples of various origins, including feces, flies, feed, dust and swabs. Surprisingly, there were no significant differences (p > 0.05) in the prevalence of resistant E. coli between the flat decks managed with conventional techniques and those managed with improved techniques. Selective cultivation delivered ampicillin- and sulfonamide-resistant E. coli proportions of up to 100% and 1.2%, respectively. While 0.5% E. coli resistant to cefotaxime and no ciprofloxacin resistance were detected. There was a significant difference (p < 0.01) in the abundance of the blaTEM-1 gene in fecal samples between EG and CG groups. The colonization of piglets with resistant pathogens before arrival, the movement of flies in the barn and the treatment of bacterial infections with antibiotics obscured the effects of hygiene improvement. Biocide tolerance tests showed no development of resistance to the farm regular disinfectant. Managing hygiene alone was insufficient for reducing antimicrobial resistances in piglet rearing. We conclude that the complex factors contributing to the presence and distribution of AMR in piglet barns underscore the necessity for a comprehensive management strategy.
Keywords: AMR; Escherichia coli; cultivation; disinfection; hygiene; pig; weaner barn.
Copyright © 2024 Jaleta, Junker, Kolte, Börger, Werner, Dolsdorf, Schwenker, Hölzel, Zentek, Amon, Nübel and Kabelitz.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Alcock B. P., Huynh W., Chalil R., Smith K. W., Raphenya A. R., Wlodarski M. A., et al. . (2023). CARD 2023: expanded curation, support for machine learning, and Resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920, PMID: - DOI - PMC - PubMed
-
- Anjum M. F., Schmitt H., Börjesson S., Berendonk T. U., Donner E., Stehling E. G., et al. . (2021). The potential of Using E. coli as an Indicator for the surveillance of antimicrobial resistance (AMR) in the environment. Curr. Opin. Microbiol. 64, 152–158. doi: 10.1016/j.mib.2021.09.011, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

