Targeting decaprenylphosphoryl-β-D-ribose 2'-epimerase for Innovative Drug Development Against Mycobacterium Tuberculosis Drug-Resistant Strains
- PMID: 38812740
- PMCID: PMC11135120
- DOI: 10.1177/11779322241257039
Targeting decaprenylphosphoryl-β-D-ribose 2'-epimerase for Innovative Drug Development Against Mycobacterium Tuberculosis Drug-Resistant Strains
Abstract
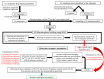
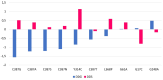
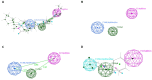
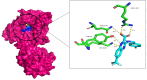
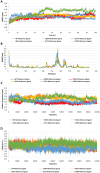
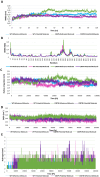
Tuberculosis (TB) remains a global health challenge with the emergence of drug-resistant Mycobacterium tuberculosis variants, necessitating innovative drug molecules. One potential target is the cell wall synthesis enzyme decaprenylphosphoryl-β-D-ribose 2'-epimerase (DprE1), crucial for virulence and survival. This study employed virtual screening of 111 Protein Data Bank (PDB) database molecules known for their inhibitory biological activity against DprE1 with known IC50 values. Six compounds, PubChem ID: 390820, 86287492, 155294899, 155522922, 162651615, and 162665075, exhibited promising attributes as drug candidates and validated against clinical trial inhibitors BTZ043, TBA-7371, PBTZ169, and OPC-167832. Concurrently, this research focused on DprE1 mutation effects using molecular dynamic simulations. Among the 10 mutations tested, C387N significantly influenced protein behavior, leading to structural alterations observed through root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), radius of gyration (Rg), and solvent-accessible surface area (SASA) analysis. Ligand 2 (ID: 390820) emerged as a promising candidate through ligand-based pharmacophore analysis, displaying enhanced binding compared with reference inhibitors. Molecular dynamic simulations highlighted ligand 2's interaction with the C387N mutation, reducing fluctuations, augmenting hydrogen bonding, and influencing solvent accessibility. These collective findings emphasize ligand 2's efficacy, particularly against severe mutations, in enhancing protein-ligand complex stability. Integrated computational and pharmacophore methodologies offer valuable insights into drug candidates and their interactions within intricate protein environments. This research lays a strategic foundation for targeted interventions against drug-resistant TB, highlighting ligand 2's potential for advanced drug development strategies.
Keywords: 3D pharmacophore; DprE1; Mycobacterium tuberculosis; resistance; virtual screening.
© The Author(s) 2024.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures
References
-
- Shirude PS, Shandil R, Sadler C, et al. Azaindoles: noncovalent DprE1 inhibitors from scaffold morphing efforts, kill Mycobacterium tuberculosis and are efficacious in vivo. J Med Chem. 2013;56:9701-9708. Accessed February 5, 2023. https://pubs.acs.org/doi/abs/10.1021/jm401382v - DOI - PubMed
LinkOut - more resources
Full Text Sources

