Spectroscopic insights into multi-phase protein crystallization in complex lysate using Raman spectroscopy and a particle-free bypass
- PMID: 38812919
- PMCID: PMC11133712
- DOI: 10.3389/fbioe.2024.1397465
Spectroscopic insights into multi-phase protein crystallization in complex lysate using Raman spectroscopy and a particle-free bypass
Abstract
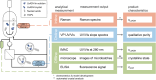
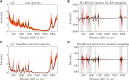
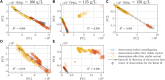
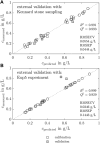
Protein crystallization as opposed to well-established chromatography processes has the benefits to reduce production costs while reaching a comparable high purity. However, monitoring crystallization processes remains a challenge as the produced crystals may interfere with analytical measurements. Especially for capturing proteins from complex feedstock containing various impurities, establishing reliable process analytical technology (PAT) to monitor protein crystallization processes can be complicated. In heterogeneous mixtures, important product characteristics can be found by multivariate analysis and chemometrics, thus contributing to the development of a thorough process understanding. In this project, an analytical set-up is established combining offline analytics, on-line ultraviolet visible light (UV/Vis) spectroscopy, and in-line Raman spectroscopy to monitor a stirred-batch crystallization process with multiple phases and species being present. As an example process, the enzyme Lactobacillus kefir alcohol dehydrogenase (LkADH) was crystallized from clarified Escherichia coli (E. coli) lysate on a 300 mL scale in five distinct experiments, with the experimental conditions changing in terms of the initial lysate solution preparation method and precipitant concentration. Since UV/Vis spectroscopy is sensitive to particles, a cross-flow filtration (cross-flow filtration)-based bypass enabled the on-line analysis of the liquid phase providing information on the lysate composition regarding the nucleic acid to protein ratio. A principal component analysis (PCA) of in situ Raman spectra supported the identification of spectra and wavenumber ranges associated with productspecific information and revealed that the experiments followed a comparable, spectral trend when crystals were present. Based on preprocessed Raman spectra, a partial least squares (PLS) regression model was optimized to monitor the target molecule concentration in real-time. The off-line sample analysis provided information on the crystal number and crystal geometry by automated image analysis as well as the concentration of LkADH and host cell proteins (HCPs) In spite of a complex lysate suspension containing scattering crystals and various impurities, it was possible to monitor the target molecule concentration in a heterogeneous, multi-phase process using spectroscopic methods. With the presented analytical set-up of off-line, particle-sensitive on-line, and in-line analyzers, a crystallization capture process can be characterized better in terms of the geometry, yield, and purity of the crystals.
Keywords: E. coli lysate; Raman spectroscopy; capture process; partial least squares regression (PLS); principal component analyses (PCA); process analytical technology (PAT); protein crystallization; ultraviolet-visible light (UV/Vis) spectroscopy.
Copyright © 2024 Wegner, Eming, Walla, Bischoff, Weuster-Botz and Hubbuch.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Raman-based PAT for VLP precipitation: systematic data diversification and preprocessing pipeline identification.Front Bioeng Biotechnol. 2024 May 31;12:1399938. doi: 10.3389/fbioe.2024.1399938. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38882637 Free PMC article.
-
In-line UV spectroscopy for the quantification of low-dose active ingredients during the manufacturing of pharmaceutical semi-solid and liquid formulations.Anal Chim Acta. 2018 Jul 12;1013:54-62. doi: 10.1016/j.aca.2018.02.007. Epub 2018 Feb 10. Anal Chim Acta. 2018. PMID: 29501092
-
Calibration-free PAT: Locating selective crystallization or precipitation sweet spot in screenings with multi-way PARAFAC models.Front Bioeng Biotechnol. 2022 Dec 14;10:1051129. doi: 10.3389/fbioe.2022.1051129. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36588941 Free PMC article.
-
Characterization of pharmaceutically relevant materials at the solid state employing chemometrics methods.J Pharm Biomed Anal. 2018 Jan 5;147:538-564. doi: 10.1016/j.jpba.2017.06.017. Epub 2017 Jun 15. J Pharm Biomed Anal. 2018. PMID: 28666554 Review.
-
Applications of process analytical technology to crystallization processes.Adv Drug Deliv Rev. 2004 Feb 23;56(3):349-69. doi: 10.1016/j.addr.2003.10.012. Adv Drug Deliv Rev. 2004. PMID: 14962586 Review.
Cited by
-
Raman-based PAT for multi-attribute monitoring during VLP recovery by dual-stage CFF: attribute-specific spectral preprocessing for model transfer.Front Bioeng Biotechnol. 2025 Aug 21;13:1631807. doi: 10.3389/fbioe.2025.1631807. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40918435 Free PMC article.
References
-
- Bakar M. R., Nagy Z. K., Rielly C. D. (2009). Seeded batch cooling crystallization with temperature cycling for the control of size uniformity and polymorphic purity of sulfathiazole crystals. Org. Process Res. Dev. 13, 1343–1356. 10.1021/op900174b - DOI
LinkOut - more resources
Full Text Sources
Research Materials

