Genetic association and transferability for urinary albumin-creatinine ratio as a marker of kidney disease in four Sub-Saharan African populations and non-continental individuals of African ancestry
- PMID: 38812969
- PMCID: PMC11134365
- DOI: 10.3389/fgene.2024.1372042
Genetic association and transferability for urinary albumin-creatinine ratio as a marker of kidney disease in four Sub-Saharan African populations and non-continental individuals of African ancestry
Abstract
Background: Genome-wide association studies (GWAS) have predominantly focused on populations of European and Asian ancestry, limiting our understanding of genetic factors influencing kidney disease in Sub-Saharan African (SSA) populations. This study presents the largest GWAS for urinary albumin-to-creatinine ratio (UACR) in SSA individuals, including 8,970 participants living in different African regions and an additional 9,705 non-resident individuals of African ancestry from the UK Biobank and African American cohorts.
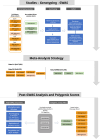
Methods: Urine biomarkers and genotype data were obtained from two SSA cohorts (AWI-Gen and ARK), and two non-resident African-ancestry studies (UK Biobank and CKD-Gen Consortium). Association testing and meta-analyses were conducted, with subsequent fine-mapping, conditional analyses, and replication studies. Polygenic scores (PGS) were assessed for transferability across populations.
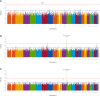
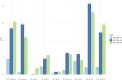
Results: Two genome-wide significant (P < 5 × 10-8) UACR-associated loci were identified, one in the BMP6 region on chromosome 6, in the meta-analysis of resident African individuals, and another in the HBB region on chromosome 11 in the meta-analysis of non-resident SSA individuals, as well as the combined meta-analysis of all studies. Replication of previous significant results confirmed associations in known UACR-associated regions, including THB53, GATM, and ARL15. PGS estimated using previous studies from European ancestry, African ancestry, and multi-ancestry cohorts exhibited limited transferability of PGS across populations, with less than 1% of observed variance explained.
Conclusion: This study contributes novel insights into the genetic architecture of kidney disease in SSA populations, emphasizing the need for conducting genetic research in diverse cohorts. The identified loci provide a foundation for future investigations into the genetic susceptibility to chronic kidney disease in underrepresented African populations Additionally, there is a need to develop integrated scores using multi-omics data and risk factors specific to the African context to improve the accuracy of predicting disease outcomes.
Keywords: African diversity; GWAS; Polygenic score; UACR; chronic kidney disease.
Copyright © 2024 Brandenburg, Chen, Boua, Govender, Agongo, Micklesfield, Sorgho, Tollman, Asiki, Mashinya, Hazelhurst, Morris, Fabian and Ramsay.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer HQ declared a past collaboration with the authors MR to the handling editor.
Figures

Update of
-
Genetic Association and Transferability for Urinary Albumin-Creatinine Ratio as a Marker of Kidney Disease in four Sub-Saharan African Populations and non-continental Individuals of African Ancestry.medRxiv [Preprint]. 2024 Apr 12:2024.01.17.24301398. doi: 10.1101/2024.01.17.24301398. medRxiv. 2024. Update in: Front Genet. 2024 May 15;15:1372042. doi: 10.3389/fgene.2024.1372042. PMID: 38293229 Free PMC article. Updated. Preprint.
Similar articles
-
Genetic Association and Transferability for Urinary Albumin-Creatinine Ratio as a Marker of Kidney Disease in four Sub-Saharan African Populations and non-continental Individuals of African Ancestry.medRxiv [Preprint]. 2024 Apr 12:2024.01.17.24301398. doi: 10.1101/2024.01.17.24301398. medRxiv. 2024. Update in: Front Genet. 2024 May 15;15:1372042. doi: 10.3389/fgene.2024.1372042. PMID: 38293229 Free PMC article. Updated. Preprint.
-
Genetic association for renal traits among participants of African ancestry reveals new loci for renal function.PLoS Genet. 2011 Sep;7(9):e1002264. doi: 10.1371/journal.pgen.1002264. Epub 2011 Sep 8. PLoS Genet. 2011. PMID: 21931561 Free PMC article.
-
Genome-wide Association Study Meta-analysis of Blood Pressure Traits and Hypertension in Sub-Saharan African Populations: An AWI-Gen Study.Res Sq [Preprint]. 2023 Feb 13:rs.3.rs-2532794. doi: 10.21203/rs.3.rs-2532794/v1. Res Sq. 2023. Update in: Nat Commun. 2023 Dec 16;14(1):8376. doi: 10.1038/s41467-023-44079-0. PMID: 36824767 Free PMC article. Updated. Preprint.
-
Interpretation of risk loci from genome-wide association studies of Alzheimer's disease.Lancet Neurol. 2020 Apr;19(4):326-335. doi: 10.1016/S1474-4422(19)30435-1. Epub 2020 Jan 24. Lancet Neurol. 2020. PMID: 31986256 Free PMC article. Review.
-
The Use of 'Omics for Diagnosing and Predicting Progression of Chronic Kidney Disease: A Scoping Review.Front Genet. 2021 Nov 8;12:682929. doi: 10.3389/fgene.2021.682929. eCollection 2021. Front Genet. 2021. PMID: 34819944 Free PMC article.
Cited by
-
A map of blood regulatory variation in South Africans enables GWAS interpretation.Nat Genet. 2025 Jul;57(7):1628-1637. doi: 10.1038/s41588-025-02223-0. Epub 2025 Jun 11. Nat Genet. 2025. PMID: 40500424 Free PMC article.
-
Cohort Profile: Africa Wits-INDEPTH partnership for Genomic studies (AWI-Gen) in four sub-Saharan African countries.Int J Epidemiol. 2024 Dec 16;54(1):dyae173. doi: 10.1093/ije/dyae173. Int J Epidemiol. 2024. PMID: 39899987 Free PMC article. No abstract available.
References
-
- Ali S. A., Soo C., Agongo G., Alberts M., Amenga-Etego L., Boua R. P., et al. (2018). Genomic and environmental risk factors for cardiometabolic diseases in Africa: methods used for Phase 1 of the AWI-Gen population cross-sectional study. Glob. Health Action 11, 1507133. 10.1080/16549716.2018.1507133 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

