Microbiological Profile of Patients with Aspiration Pneumonia Identified by Combined Detection Methods
- PMID: 38813526
- PMCID: PMC11135560
- DOI: 10.2147/IDR.S461935
Microbiological Profile of Patients with Aspiration Pneumonia Identified by Combined Detection Methods
Abstract
Purpose: Aspiration pneumonia (AP) challenges public health globally. The primary aim of this study was to ascertain the microbiological profile characteristics of patients with AP evaluated by combined detection methods, including conventional microbiological tests (CMTs), chips for complicated infection detection (CCID), and metagenomic next-generation sequencing (mNGS).
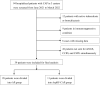
Patients and methods: From June 2021 to March 2022, a total of thirty-nine patients with AP or community-acquired pneumonia with aspiration risk factors (AspRF-CAP) from 3 hospitals were included. Respiratory specimens, including bronchoalveolar lavage fluid (BALF), sputum, and tracheal aspirate, were collected for microorganism detection.
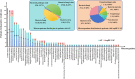
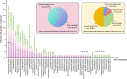
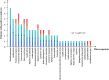
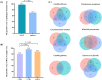
Results: Patients with AP were more inclined to be older, to have a shorter duration from illness onset to admission, to have a higher prevalence of different underlying diseases, particularly diabetes mellitus, chronic heart disease, and cerebrovascular disease, and to have a higher CURB-65 score (all P < 0.05). A total of 213 and 31 strains of microorganisms were detected in patients with AP and AspRF-CAP, respectively. The most common pathogens in AP were Corynebacterium striatum (17/213, 7.98%), Pseudomonas aeruginosa (15/213, 7.04%), Klebsiella pneumoniae (15/213, 7.04%), and Candida albicans (14/213, 6.57%). Besides, the most common pathogens in AspRF-CAP were Candida albicans (5/31, 16.13%), Pseudomonas aeruginosa (3/31, 9.68%) and Klebsiella pneumoniae (3/31, 9.68%). Moreover, Klebsiella pneumoniae (7/67, 10.45%) and Candida glabrata (5/67, 7.46%) were the most common pathogens among the 9 non-survived patients with AP.
Conclusion: The prevalent pathogens detected in cases of AP were Corynebacterium striatum, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Candida albicans. Early combined detection methods for patients with AP enhance the positive detection rate of pathogens and potentially expedites the initiation of appropriate antibiotic therapeutic strategies.
Keywords: aspiration; chips; combined detection; metagenomic next-generation sequencing; microbiology; pneumonia.
© 2024 Xu et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures
Similar articles
-
Diagnostic value of bronchoalveolar lavage fluid metagenomic next-generation sequencing in pediatric pneumonia.Front Cell Infect Microbiol. 2022 Oct 27;12:950531. doi: 10.3389/fcimb.2022.950531. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36389175 Free PMC article.
-
Clinical Efficiency of Metagenomic Next-Generation Sequencing in Sputum for Pathogen Detection of Patients with Pneumonia According to Disease Severity and Host Immune Status.Infect Drug Resist. 2023 Sep 6;16:5869-5885. doi: 10.2147/IDR.S419892. eCollection 2023. Infect Drug Resist. 2023. PMID: 37700802 Free PMC article.
-
Diagnostic value of metagenomic next-generation sequencing of bronchoalveolar lavage fluid for the diagnosis of suspected pneumonia in immunocompromised patients.BMC Infect Dis. 2022 Apr 29;22(1):416. doi: 10.1186/s12879-022-07381-8. BMC Infect Dis. 2022. PMID: 35488253 Free PMC article.
-
[Diagnostic value of detection of pathogens in bronchoalveolar lavage fluid by metagenomics next-generation sequencing in organ transplant patients with pulmonary infection].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2021 Dec;33(12):1440-1446. doi: 10.3760/cma.j.cn121430-20211008-01439. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2021. PMID: 35131010 Chinese.
-
Microbiological diagnostic performance of metagenomic next-generation sequencing compared with conventional culture for patients with community-acquired pneumonia.Front Cell Infect Microbiol. 2023 Mar 16;13:1136588. doi: 10.3389/fcimb.2023.1136588. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37009509 Free PMC article.
Cited by
-
Rising prevalence and drug resistance of Corynebacterium striatum in lower respiratory tract infections.Front Cell Infect Microbiol. 2025 Jan 7;14:1526312. doi: 10.3389/fcimb.2024.1526312. eCollection 2024. Front Cell Infect Microbiol. 2025. PMID: 39839260 Free PMC article. Review.
-
High co-infection burden and ICU-specific pathogen profiles in pediatric Candida albicans respiratory infections: a large-scale tNGS analysis.Eur J Clin Microbiol Infect Dis. 2025 Jul 18. doi: 10.1007/s10096-025-05216-3. Online ahead of print. Eur J Clin Microbiol Infect Dis. 2025. PMID: 40679578
-
Dentures and the oral microbiome: Unraveling the hidden impact on edentulous and partially edentulous patients - a systematic review and meta-analysis.Evid Based Dent. 2025 May 29. doi: 10.1038/s41432-025-01149-0. Online ahead of print. Evid Based Dent. 2025. PMID: 40442493
References
LinkOut - more resources
Full Text Sources
Miscellaneous

