Gene signatures of endoplasmic reticulum stress and mitophagy for prognostic risk prediction in lung adenocarcinoma
- PMID: 38813617
- PMCID: PMC11179159
- DOI: 10.1049/syb2.12092
Gene signatures of endoplasmic reticulum stress and mitophagy for prognostic risk prediction in lung adenocarcinoma
Abstract
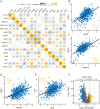
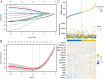
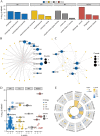
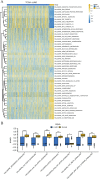
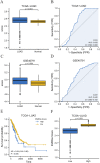
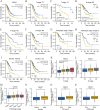
Genes associated with endoplasmic reticulum stress (ERS) and mitophagy can be conducive to predicting solid tumour prognosis. The authors aimed to develop a prognosis prediction model for these genes in lung adenocarcinoma (LUAD). Relevant gene expression and clinical information were collected from public databases including Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA). A total of 265 differentially expressed genes was finally selected (71 up-regulated and 194 downregulated) in the LUAD dataset. Among these, 15 candidate ERS and mitophagy genes (ATG12, CSNK2A1, MAP1LC3A, MAP1LC3B, MFN2, PGAM5, PINK1, RPS27A, SQSTM1, SRC, UBA52, UBB, UBC, ULK1, and VDAC1) might be critical to LUAD based on the expression analysis after crossing with the ERS and mitochondrial autophagy genes. The prediction model demonstrated the ability to effectively predict the 5-, 3-, and 1-year prognoses of LUAD patients in both GEO and TCGA databases. Moreover, high VDAC1 expression was associated with poor overall survival in LUAD (p < 0.001), suggesting it might be a critical gene for LUAD prognosis prediction. Overall, the prognosis model based on ERS and mitophagy genes in LUAD can be useful for evaluating the prognosis of patients with LUAD, and VDAC1 may serve as a promising biomarker for LUAD prognosis.
Keywords: bioinformatics; genomics; lung; tumours.
© 2024 The Authors. IET Systems Biology published by John Wiley & Sons Ltd on behalf of The Institution of Engineering and Technology.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Construction and validation of a prognostic model for lung adenocarcinoma based on endoplasmic reticulum stress-related genes.Sci Rep. 2022 Nov 18;12(1):19857. doi: 10.1038/s41598-022-23852-z. Sci Rep. 2022. PMID: 36400857 Free PMC article.
-
Building endoplasmic reticulum stress-related LncRNAs signatures of lung adenocarcinoma.J Gene Med. 2024 Aug;26(8):e3731. doi: 10.1002/jgm.3731. J Gene Med. 2024. PMID: 39146558
-
Characterization of an endoplasmic reticulum stress-associated lncRNA prognostic signature and the tumor-suppressive role of RP11-295G20.2 knockdown in lung adenocarcinoma.Sci Rep. 2024 May 29;14(1):12283. doi: 10.1038/s41598-024-62836-z. Sci Rep. 2024. PMID: 38811828 Free PMC article.
-
Prediction of prognosis and immune response in lung adenocarcinoma based on mitophagy and lactate-related gene signatures.Int J Clin Oncol. 2025 Feb;30(2):277-297. doi: 10.1007/s10147-024-02664-3. Epub 2024 Nov 27. Int J Clin Oncol. 2025. PMID: 39601968
-
The role and machine learning analysis of mitochondrial autophagy-related gene expression in lung adenocarcinoma.Front Immunol. 2025 Apr 17;16:1509315. doi: 10.3389/fimmu.2025.1509315. eCollection 2025. Front Immunol. 2025. PMID: 40313958 Free PMC article.
Cited by
-
α-asarone activates mitophagy to relieve diabetic encephalopathy via inhibiting apoptosis and oxidative stress.Metab Brain Dis. 2025 Feb 15;40(2):126. doi: 10.1007/s11011-025-01556-3. Metab Brain Dis. 2025. PMID: 39954135
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

