LILRB4 represents a promising target for immunotherapy by dual targeting tumor cells and myeloid-derived suppressive cells in multiple myeloma
- PMID: 38813706
- PMCID: PMC11532705
- DOI: 10.3324/haematol.2024.285099
LILRB4 represents a promising target for immunotherapy by dual targeting tumor cells and myeloid-derived suppressive cells in multiple myeloma
Abstract
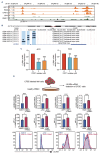
Multiple myeloma (MM) remains an incurable hematologic malignancy. Despite tremendous advances in the treatment of this disease, about 10% of patients still have very poor outcomes with a median overall survival of less than 24 months. Our study aimed to underscore the critical mechanisms pertaining to rapid disease progression and provide novel therapeutic choices for these ultrahigh-risk patients. We utilized single-cell transcriptomic sequencing to dissect the characteristic bone marrow niche of patients who survived less than 2 years (EM24). Notably, enrichment of a LILRB4high pre-mature plasma-cell cluster was observed in EM24 patients compared to patients with durable remission. This cluster exhibited aggressive proliferation and a drug-resistance phenotype. High levels of LILRB4 promoted MM clonogenicity and progression. Clinically, high expression of LILRB4 was correlated with poor prognosis in both newly diagnosed MM patients and relapsed/ refractory MM patients. ATAC-sequencing analysis identified that pronounced chromosomal accessibility caused the elevation of LILRB4 on MM cells. CRISPR-Cas9 deletion of LILRB4 alleviated the growth of MM cells, inhibited the immunosuppressive function of myeloid-derived suppressive cells (MDSC), and further rescued T-cell dysfunction in the MM microenvironment. Greater infiltration of MDSC was observed in EM24 patients. We therefore generated an innovative T-cell receptor-based chimeric antigen receptor T cell, LILRB4-STAR-T. Cytotoxicity experiments demonstrated that LILRB4-STAR-T cells efficaciously eliminated tumor cells and impeded MDSC function. In conclusion, our study elucidates that LILRB4 is an ideal biomarker and promising immunotherapy target for high-risk MM. LILRB4-STAR-T-cell immunotherapy is promising against both tumor cells and the immunosuppressive tumor microenvironment in MM.
Figures







References
-
- Bazarbachi AH, Al Hamed R, Malard F, et al. . Relapsed refractory multiple myeloma: a comprehensive overview. Leukemia. 2019;33(10):2343-2357. - PubMed
-
- Cowan AJ, Green DJ, Kwok M, et al. . Diagnosis and management of multiple myeloma: a review. JAMA. 2022;327(5):464-477. - PubMed
-
- Ríos-Tamayo R, Sáinz J, Martínez-López J, et al. . Early mortality in multiple myeloma: the time-dependent impact of comorbidity: a population-based study in 621 real-life patients. Am J Hematol. 2016;91(7):700-704. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

