Functional analysis of epilepsy-associated GABAA receptor mutations using Caenorhabditis elegans
- PMID: 38813985
- PMCID: PMC11296113
- DOI: 10.1002/epi4.12982
Functional analysis of epilepsy-associated GABAA receptor mutations using Caenorhabditis elegans
Abstract
Objective: GABAA receptor subunit mutations pose a significant risk for genetic generalized epilepsy; however, there are over 150 identified variants, many with unknown or unvalidated pathogenicity. We aimed to develop in vivo models for testing GABAA receptor variants using the model organism, Caenorhabditis elegans.
Methods: CRISPR-Cas9 gene editing was used to create a complete deletion of unc-49, a C. elegans GABAA receptor, and to create homozygous epilepsy-associated mutations in the endogenous unc-49 gene. The unc-49 deletion strain was rescued with transgenes for either the C. elegans unc-49B subunit or the α1, β3, and γ2 subunits for the human GABAA receptor. All newly created strains were analyzed for phenotype and compared against existing unc-49 mutations.
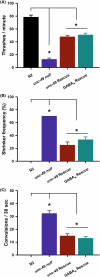
Results: Nematodes with a full genetic deletion of the entire unc-49 locus were compared with existing unc-49 mutations in three separate phenotypic assays-coordinated locomotion, shrinker frequency and seizure-like convulsions. The full unc-49 deletion exhibited reduced locomotion and increased shrinker frequency and PTZ-induced convulsions, but were not found to be phenotypically stronger than existing unc-49 mutations. Rescue with the unc-49B subunit or creation of humanized worms for the GABAA receptor both showed partial phenotypic rescue for all three phenotypes investigated. Finally, two epilepsy-associated variants were analyzed and deemed to be loss of function, thus validating their pathogenicity.
Significance: These findings establish C. elegans as a genetic model to investigate GABAA receptor mutations and delineate a platform for validating associated variants in any epilepsy-associated gene.
Plain language summary: Epilepsy is a complex human disease that can be caused by mutations in specific genes. Many possible mutations have been identified, but it is unknown for most of them whether they cause the disease. We tested the role of mutations in one specific gene using a small microscopic worm as an animal model. Our results establish this worm as a model for epilepsy and confirm that the two unknown mutations are likely to cause the disease.
Keywords: convulsion; locomotion; seizure; shrinker; unc‐49.
© 2024 The Author(s). Epilepsia Open published by Wiley Periodicals LLC on behalf of International League Against Epilepsy.
Conflict of interest statement
None of the authors has any conflict of interest to disclose. We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Figures



References
-
- Perucca P, Bahlo M, Berkovic SF. The genetics of epilepsy. Annu Rev Genomics Hum Genet. 2020;21:205–230. - PubMed
-
- Ellis CA, Petrovski S, Berkovic SF. Epilepsy genetics: clinical impacts and biological insights. Lancet Neurol. 2020;19:93–100. - PubMed
-
- Cunliffe VT. Building a zebrafish toolkit for investigating the pathobiology of epilepsy and identifying new treatments for epileptic seizures. J Neurosci Methods. 2016;260:91–95. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

